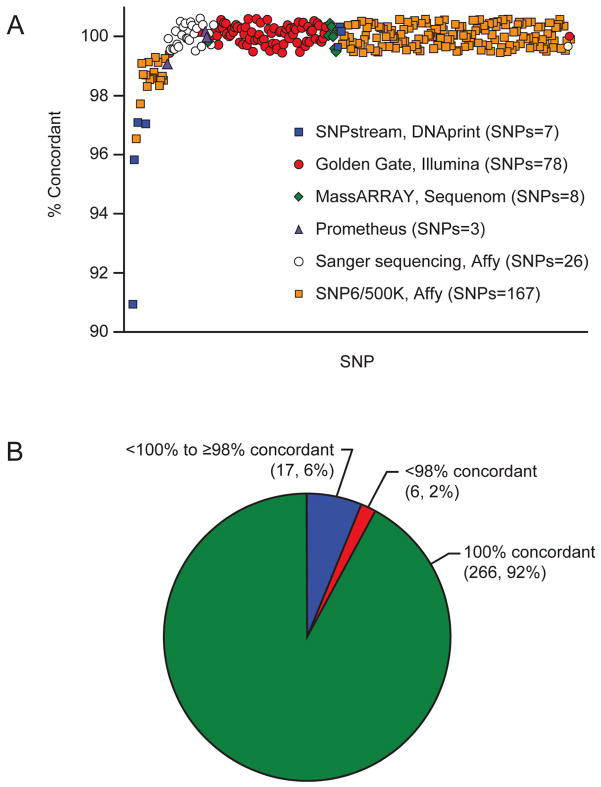
Figure 3. Concordance of the DMET Plus array with other genotyping methods.
The DMET genotyping was compared to all other sequencing and genotyping data available in 220 patients. A total of 259 SNPs were assessed by the DMET array and at least one orthogonal method. Of these, 233 SNPS were assessed by one orthogonal method, 22 SNPs were assessed by two orthogonal methods, and 4 SNPs were assessed by three orthogonal methods, yielding a total of 289 tests of concordance between an orthogonal method and a DMET SNP call. (A) 167 DMET SNPs were in common with the Affymetrix Gene Chip (orange squares), 3 SNPs with Prometheus TPMT genotyping (purple triangles), 78 with the Illumina Golden Gate chip (red circles), 8 with the Sequenom iplex Gold MassARRAY platform (green diamonds), 26 with Sanger sequencing (white circles), and 7 with Beckman Coulter GenomeLab SNPstream (blue squares). (B) A threshold of 98% was used to determine SNPs with poor concordance. SNPs below this threshold (6, 2%) were further investigated, where possible, by comparing the discordant genotypes with any other overlapping genotyping methods.