Figure 4 .
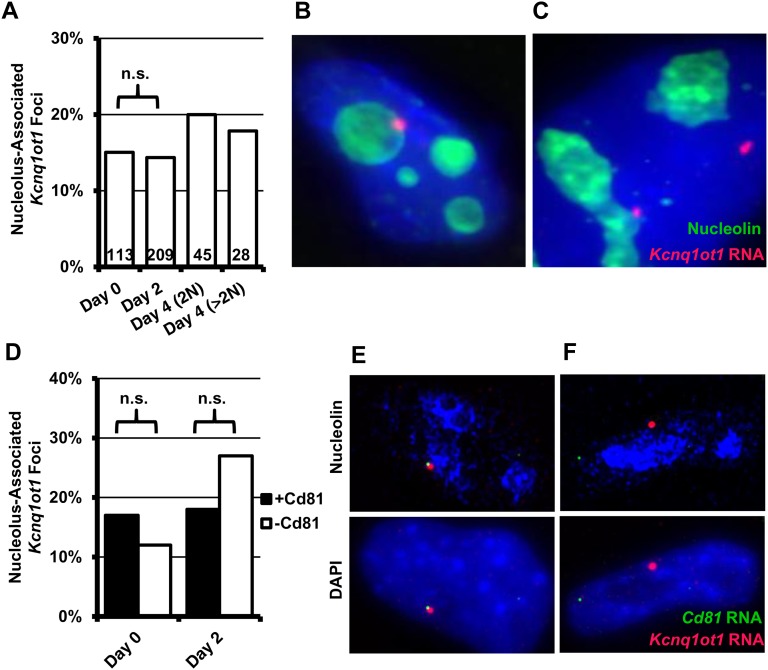
Localization of Kcnq1ot1 RNA foci and active paternal Cd81 in Eed−/− TS cells. (A) Summary of Kcnq1ot1 RNA localization in undifferentiated and differentiated Eed−/− TS cells. Representative images (Z-stack projections) of undifferentiated TS (B) and endoreduplicated (C) Eed-null cells (DAPI, blue; Nucleolin, green; Kcnq1ot1 ncRNA, red). (D) To determine whether paternal expression of placentally imprinted genes was compatible with nucleolar association, we compared localization frequency of Kcnq1ot1 RNA foci overlapping Cd81 RNA FISH signals (black bars) against Kcnq1ot1 foci without detectable Cd81 (open bars) in undifferentiated and differentiated Eed−/− TS cells. Nucleolar association of paternal Cd81 RNA FISH signals in Eed−/− TS was not statistically different from localization of Kcnq1ot1 RNA foci. Representative images of cells with overlapping Kcnq1ot1 (red) and Cd81 (green) RNA FISH signals, which we counted as +Cd81, (E) and without, which were counted as −Cd81 (F). Top shows RNA FISH with nucleolin immunofluorescence; bottom with DAPI staining. For panels (A) and (D), the total number of RNA FISH foci analyzed (n) are shown within each bar. Statistical significance was determined by χ-squared tests; n.s., not significant.