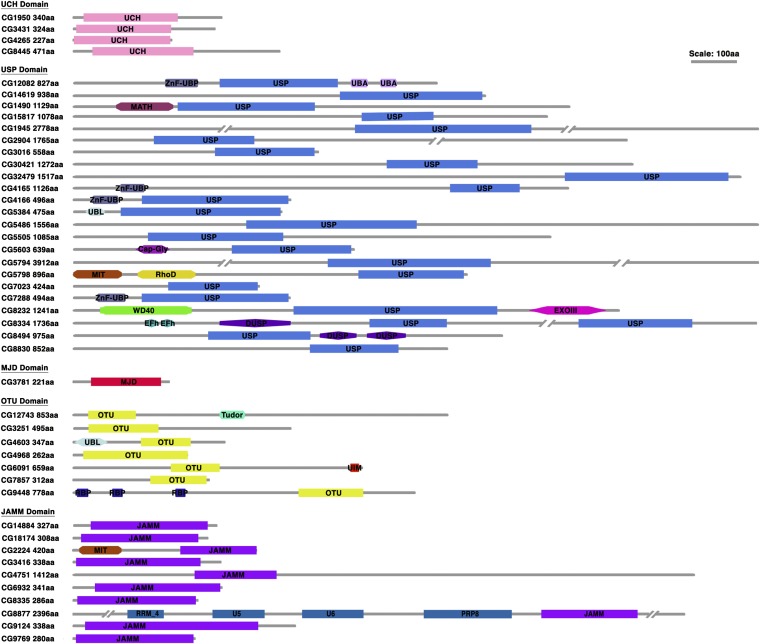
Figure 1 .
Inventory and domain architectures of annotated DUBs in Drosophila. Drosophila DUBs are characterized into five subfamilies on the basis of their signature DUB catalytic domains. The UCH, USP, MJD, OUT, and JAMM domain-containing proteases are shown. Apart from signature DUB domains, we retrieved domain architectures for each DUB by using the Pfam and the NCBI Conserved Domain database. The abbreviations for additional domains are listed as follows: Cap-Gly, cytoskeleton-associated proteins, glycine-rich domain; DUSP, domain in ubiquitin-specific proteases; EFh, EF-hand, calcium binding motif; EXOIII, exonuclease, RNase T/DNA polymerase III; MATH, meprin and TRAF-homology; MIT, microtubule interacting and trafficking molecule domain; PRP8, pre-mRNA processing splicing factor 8; RBP, zinc finger, RanBP2-type; RhoD, rhodanese homology domain; RPT, internal repeats; RRM_4, RNA recognition motif of the spliceosomal PrP8; Tudor, Tudor domain; U5 and U6, U5 and U6 snRNA binding domains; UBA, ubiquitin-associated; UBL, ubiquitin-like; UIM, ubiquitin interaction motif; WD40, WD40-repeat-containing domain; and ZnF-UBP, zinc finger ubiquitin binding domain. Proteins and domains are plotted on an approximate scale.