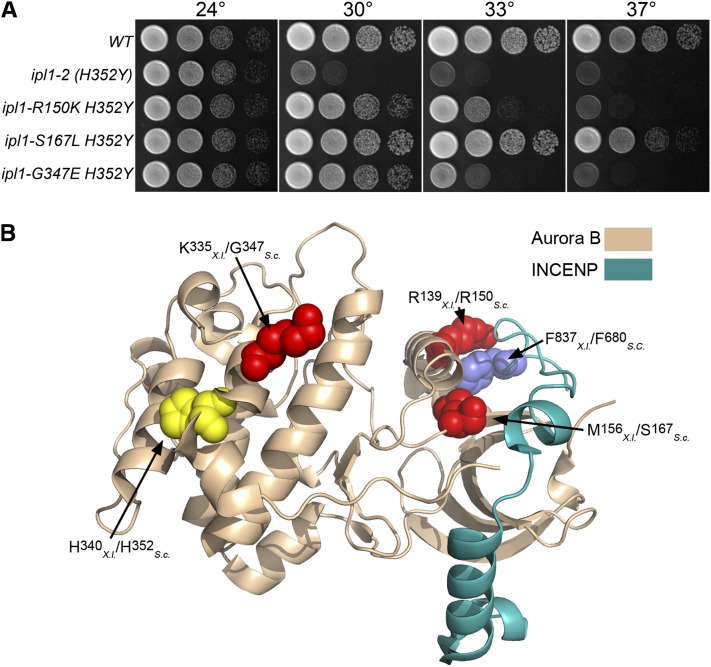
Figure 1 .
Intragenic ipl1-2 suppressors. (A) Cultures of WT (KT1112), ipl1-2 (H352Y) (KT1829), ipl1-2 R150K H352Y (KT2865), ipl1-S167L H352Y (KT2867), and ipl1-G347E H352Y (KT2869) strains were serially diluted onto YPD medium and imaged after 40 hr at the designated temperatures. (B) The locations of intragenic ipl1-2 suppressor mutations are mapped on the X. laevis Aurora B- INCENP structure [2BFX.pdb (Sessa et al. 2005)]. The highlighted amino acid residues are represented as space-filling models. The location of the ipl1-2 mutation H352Y is shown in yellow. Residues altered by intragenic suppressor mutations are red, and the amino acid residue in INCENP that associates with R139 is blue. Note that both S167 and R150 are predicted to lie near the interface with INCENP/Sli15.