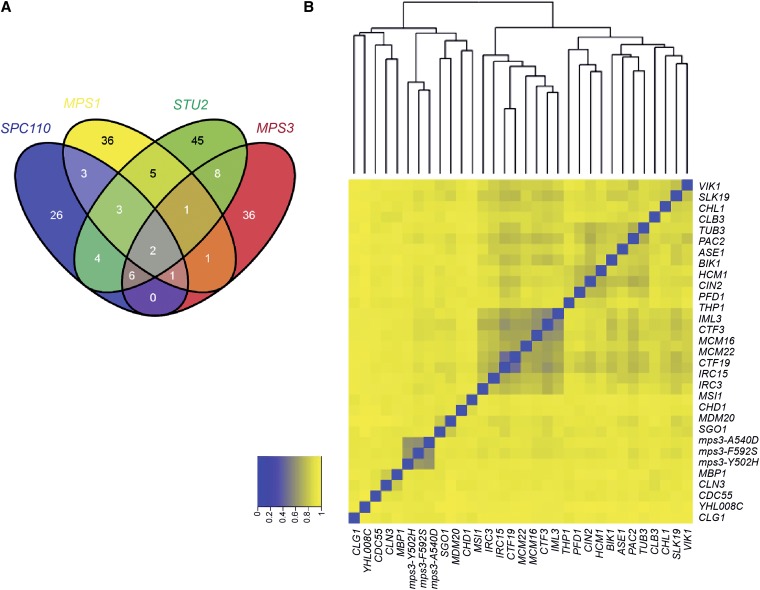
Figure 4 .
Similarity of mps3 SUN domain mutants to other cell cycle regulators. (A) Venn diagram showing overlap of synthetic lethal, synthetic sick or negative genetic interactions identified in analyses of spc110, mps1, stu2, and mps3-SUN domain mutants. (B) Synthetic lethal, synthetic sick, or negative genetic interactions were extracted for yeast gene pairs from BioGRID 3.1.88 and integrated with our data. To characterize the similarity of interactions found with mps3-SUN domain mutants to interactions identified with other deletions, we used the Jaccard distance, which takes into account the intersection and union of datasets (Jay et al. 2012). A total of 32 genes have a Jaccard distance <0.95 to all three mps3-SUN mutants, indicating a similar genetic signature. Genes were organized using two-dimensional hierarchical clustering based on their Jaccard distance from one another. Blue, Jaccard index = 0, which is very similar; yellow, Jaccard index = 1, less similar.