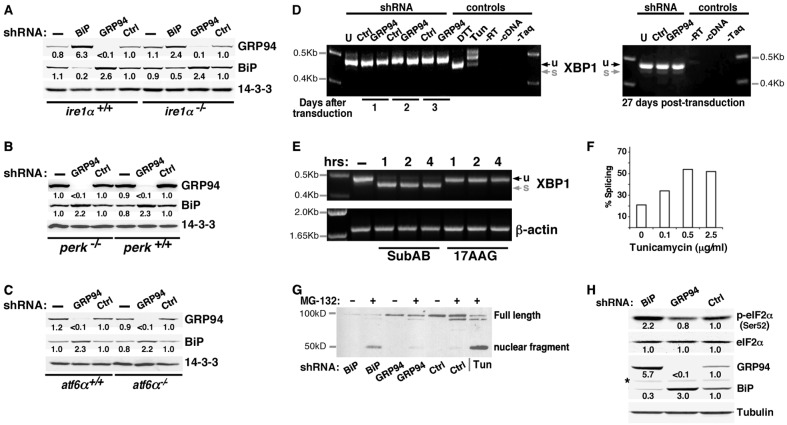
Fig. 4.
The response to GRP94 ablation is not mediated by UPR signaling. (A–C) The GRP94-specific response is observed in MEFs deficient for each of the three UPR transducers. (A) GRP94 and BiP were silenced individually, using lentiviral infections, in IRE1-deficient MEFs. Cell lysates were analyzed by western blotting. Relative band intensities, normalized to the loading control (14-3-3) are indicated. (B) Response of PERK-deficient MEFs to lentiviral-mediated silencing of GRP94. (C) Response of ATF6α-deficient MEFs to lentiviral-mediated silencing of GRP94. (D) XBP1 splicing is not triggered in GRP94-depleted cells. Total RNA was extracted from 293T cells that were either untransduced (U), transduced with a lentivirus with shRNA to GRP94 (GRP94) or with an irrelevant shRNA (Ctrl). Cells were harvested at the indicated times after viral transduction and XBP1 was amplified by RT-PCR. The unspliced (u, 473 bp) and spliced (s, 447 bp) forms of XBP1 differ by 26 nucleotides. Only the unspliced form is seen in all samples, whereas the positive controls for cells mounting active UPR show either efficient splicing (DTT treatment, 1 µM for 6 hours) or partial splicing (Tun, 10 µg/ml for 8 hours). (E) Inhibition of GRP94 does not trigger XBP1 splicing, whereas ablation of BiP does. XBP1 splicing in 10T1/2 cells that were treated with 100 ng/ml subAB, a toxin that cleaves BiP selectively and induces global UPR, or exposed to 10 µM 17AAG overnight. Cells were harvested at the indicated time points. RNA extracts were assayed as in panel D, with β-actin amplification serving as a loading control. (F) Sensitivity of detection of XBP1 splicing. 293T cells were subjected to the indicated doses of tunicamycin overnight and splicing was detected as shown in Fig. 4D,E. The percentage splicing is the ratio of the spliced band over the total amplicon [s/(u + s)] in each gel lane. Even though there is a higher level of basal activity in our 3T3 cells compared with that of the 293T cells or MEFs, splicing in response to as little as 0.1 µg/ml tunicamcyin was already detectable. Doses up to 2.0 µg/ml were used in the other figures. (G) ATF6 endo-proteolysis is induced in BiP-, but not in GRP94-depleted cells. Nuclear extracts of 293T cells stably co-expressing HA-tagged ATF6 with shRNA against BiP, GRP94 or Ctrl genes were analyzed by immunoblotting. The nuclear ATF6 fragment was detected with anti-HA antibody. Consistent with previous results (Haze et al., 1999), the nuclear ATF6 fragment was detected when cells were treated with tunicamycin (Tun; 2 µg/ml for 3 hours) to induce ER stress. The same fragment is induced in BiP-depleted cells, whereas the pattern of GRP94-depleted is similar to the control cells. Enhanced levels of the ATF6 fragment were detected when cells were treated with MG132 (10 µM for 3 hours) to prevent degradation. A longer exposure is presented in supplementary material Fig. S4C. (H) Depletion of BiP, but not of GRP94, triggers PERK activity. BiP, GRP94 or an irrelevant gene (Ctrl) were silenced with the appropriate shRNA-encoding lentivirus and then assayed by western blotting with the indicated antibodies. The anti-phospho-eIF2a antibody measures the phosphorylation site most indicative of PERK activation. The asterisk indicates a non-specific band. The data shown in all panels of this figure are representative of at least three independent experiments.