Genetically expressed fluorescent proteins (FPs)[1–3] revolutionized the field of cell biology, especially enabling live-cell studies.[4–16] The limitations of FPs however are related to the photophysical properties of their fluorophores, mainly brightness and photostability, which are still are inferior to organic fluorophores.[17–19] Hybrid tagging strategies, such as SNAP-tag[20–22] and HaloTag[23], as well as genetic encoding of fluorescent amino acids[24] are alternatives to FPs that allow genetic targeting of synthetic dyes.[25] Fluorogen activating proteins (FAPs) belong to a recently developed hybrid-tagging strategy that allows target-specific activation of the fluorescence of otherwise nonfluorescent dyes (fluorogens)[26] by hundreds-to-thousands fold. FAPs that bind derivatives of Malachite Green (MG) displayed the greatest fluorogenic activation, up to 20000-fold, primarily because of an extremely low unbound dye fluorescence in water.[26] The high specificity and extremely low background signal from FAP tagging were shown to be very useful in fluorescence microscopy of live cells[27] including super-resolution STED imaging.[28]
Recently we developed an efficient approach for the preparation of fluorogenic dyes with significantly improved brightness.[29] In a strategy derived from significant historical work on light-harvesting polymers[30–32], this approach is based on the decoration of a single MG fluorogen with one or more dyes that have a high extinction coefficient, for example, Cy3, at a short enough distance to undergo efficient intramolecular Forster Resonance Energy Transfer (FRET). As a result a tandem such as Cy3-MG dye possesses brightness under the Cy3 excitation that is two-fold higher than the inherent brightness of the MG-FAP complex. Increasing the number of donors increased the molecular brightness in step with the number of donors. The extinction coefficients achieved with 4 donors are up to 5×105 M−1cm−1, approaching quantum dot extinction coefficients. The genetically expressed protein is ~25 kDa, and the multi chromophore structure is 3 kDa, an overall fluorescent complex about the size of GFP, but nearly a factor of 10 brighter.[29]
These Cy3-MG structures, however bright, have some limitations as live-cell labels: 1) due to the presence of multiple negative charges these dyes do not penetrate through the plasma membrane of living cells and as result have limited application for intracellular studies; 2) while the ability to monitor non-bound dye may be useful for many studies, Cy3-MG tandems (similarly to MG) are practically non-fluorescent in the absence of FAP; 3) they have low brightness under 2-photon irradiation that limits their application in more complex tissue experiments where the bright fluorogenic properties could be highly valuable.
In this work we present tetramethyl rhodamine-MG tandem dyes (Scheme 1) which are designed to overcome these shortcomings. These cationic dyes penetrate through the plasma membrane of live cells and have photophysical properties suitable for 2-photon imaging. Surprisingly, the TMR-para-MG dye with a 3-carbon linker displays colour-switching properties upon binding the FAP, rather than the fluorogenic activation that was seen with Cy3-MG and is shown here for TMR-meta-MG tandem dye. This finding provides a new class of targeting and fluorescence modification that can potentially be exploited in the design of genetically encoded biosensors using fluorogen activating peptides.
Scheme 1.
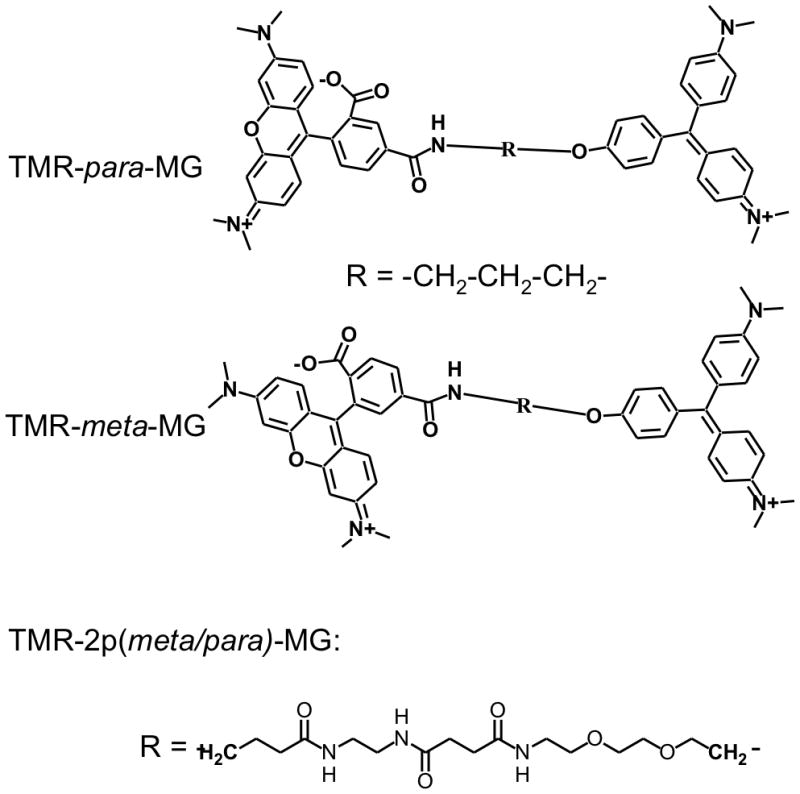
Chemical structures of TMR-MG tandem dyes.
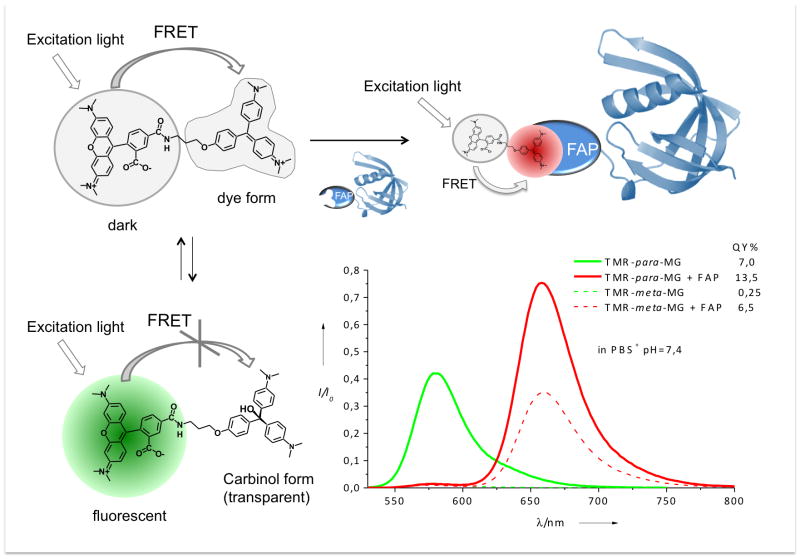
The TMR-MG dyes were designed for efficient energy transfer from TMR to MG (SI Figure S10). As result these dyes have a strong fluorescence of the MG fluorophore with an emission maximum ~660 nm in the presence of FAP and virtually complete quenching of the TMR fluorophore (Figure 1). In the absence of FAP, the TMR-para-MG dye is characterized by a pronounced fluorescence of the TMR fluorophore with emission maximum at 580 nm (Figure 1). This switch in the emission spectrum of TMR-MG upon binding to the FAP is due to the chemical properties of MG. The MG dye exists in equilibrium between two forms at physiological pHs: the dye form which is characterized by strong absorbance with maximum at 609 nm and the decolourized carbinol form (SI Figure S1.5).
Figure 1.
Schematic representation of dual-colour behaviour of TMR-para-MG dye and emission spectra of TMR-para-MG (and TMR-meta-MG, dashed lines) in the absence and in the presence of FAP. Fluorescence spectra are normalized to absorbance at 515 nm, excitation 515 nm.
The pKa of MG-NH2, the precursor of TMR-MG, is close to 7.7 (SI Figure S16) and at pH 7.4 approximately 35% of this compound exists in carbinol form (cMG-NH2). MG retains these chemical properties after coupling to TMR in the TMR-para-MG dye (Figure 2b), but it has a slightly higher pKa (8.0). As result in TMR-para-MG at pH 7.4, ~70% of the TMR fluorescence is quenched due to FRET to the MG acceptor, which is not fluorescent in the absence of FAP. The remaining unquenched TMR donors (30%) are responsible for the emission of TMR-para-MG when the dye is not bound to FAP (Figure 2, 4b). In the presence of FAP only TMR-MG binds and the TMR-cMG does not bind until it is converted to the TMR-MG form. For this reason the TMR-MG-FAP complex displays efficient energy transfer from TMR to MG and is characterized by pronounced energy transfer-sensitized fluorescence emission from the MG fluorophore and quenched TMR fluorescence (Figure 1).
Figure 2.
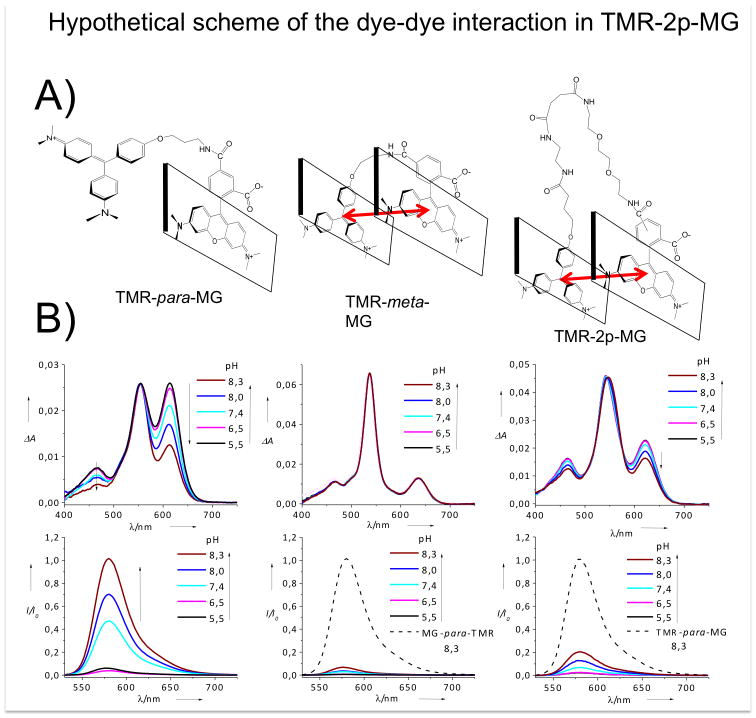
A) Hypothetical scheme of the chromophore-chromophore interaction in TMR-para-MG, TMR-meta-MG and TMR-2p-MG dyes; B) Absorbance and normalized fluorescence spectra of TMR-para-MG, TMR-meta-MG and TMR-2p(para)-MG dyes in phosphate buffer at different pH.
We found that the colour-switching behaviour of the TMR-MG dye requires a short linker between the two chromophores, preventing the intramolecular heterochromophore interaction. When permitted, this interaction leads to quenching of the TMR fluorescence, as is evident in the meta isomer (Figures 2, 3). The interaction between MG and TMR fluorophores in TMR-meta-MG dye is also confirmed by absorbance and NMR data (SI). In aqueous solutions TMR-meta-MG displays a ~15 nm hypsochromic-shift in the TMR absorbance and a ~30 nm bathochromic shift in the MG absorbance, along with a change in the relative absorbance of these spectral components (Figure 2b, SI Figure S11, Table S1). These spectroscopic changes are consistent with formation of an intramolecular hetero-H-aggregate through pi-pi stacking of the MG and TMR chromophores.[33] In this case, the antisymmetric H-aggregate dipole is not reduced to zero because the MG and TMR dipole moments are not of equal magnitude. Unlike MG and TMR-para-MG dyes TMR-meta-MG shows no response from pH 6—8 (Figure 2b) decolorizing only at high pH (SI Figure S19). Interaction of TMR-meta-MG with FAP also leads to the disruption of the heterochromophore complex but this is much slower process than binding of MG or TMR-para-MG to the same FAP under identical conditions (SI Figure S17). Elongation of the linker between MG and TMR fluorophores leads to a pair of isomeric TMR-2p(meta/para)-MG dyes with indistinguishable spectroscopic and very similar chemical properties (SI Figure S6, Table S1, Figure S11–S12). Their absorbance and fluorescence spectra are intermediate between TMR-meta-MG and TMR-para-MG dyes. They display much lower pH sensitivity as compared to TMR-para-MG, but somewhat higher as compared to TMR-meta-MG dye (Figure 2c, SI). This is likely due to the intramolecular heterochromophore interaction between MG and TMR in TMR-2p(meta/para)-MG dyes analogously to TMR-meta-MG. However this interaction is not as pronounced as in the case of the 3-carbon linked TMR-meta-MG.
Figure 3.
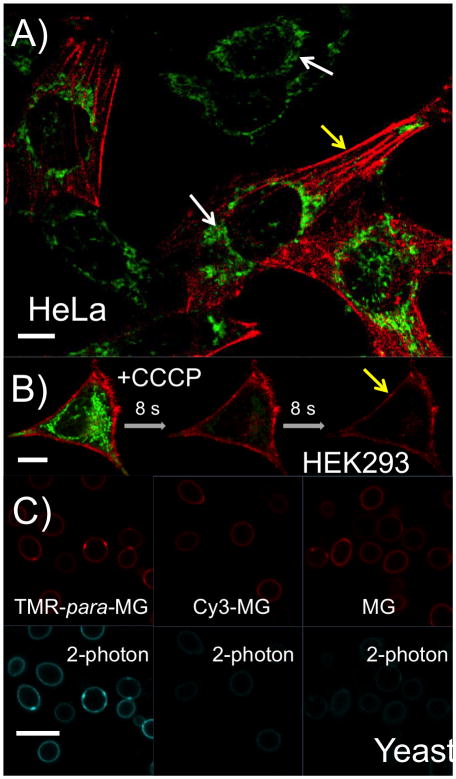
A) Live-cell confocal microscopy images of HeLa cells that express FAP dH6 fused to the filament forming protein actin after incubation with 150 nM TMR-para-MG dye for 30 min. Green channel corresponds to emission of TMR (575–630 nm) excited with 561 nm laser; red channel corresponds to emission of MG bound to FAP (650–710 nm) excited with 633 nm laser; B) Time series of live-cell spinning-disc images of the HEK293 cells that express dL5 E52D L91S fused to the transmembrane domain of B7-1 protein for inner-leaflet display in the presence of 150 nM TMR-para-MG after treatment with 100 μM CCCP. Green: TMR channel; Red: MG channel; C) Confocal images of live yeast cells with surface-displayed dL5 E52D L91S in the presence of 150 nM TMR-para-MG, Cy3-MG and MG dyes correspondingly. Red: MG channel; Blue: 2-photon channel, excitation is 850 nm and emission is 650–710 nm. Scale bar is 10 μm.
Because of their positive charge TMR-MG dyes can penetrate through the plasma membrane of living cells. Similar to other cationic dyes, such as TMRM[34], they predominantly accumulate in mitochondria (SI Figure S24, Movie S1). Due to such accumulation and the relatively high pH in mitochondria (~8.0)[35], TMR-para-MG stains these organelles well (Figure 3a, b) with pronounced fluorescence in the TMR channel.
One advantage of TMR-para-MG dye is its ability to distinguish the cells that express FAP from the cells that do not express FAP (Figure 3a). In this virally transduced population, some cells in the presence of TMR-para-MG show only TMR signal (in green, white arrows) while other cells that do express FAP show both TMR fluorescence from mitochondria and site-specific fluorogen labelling with the MG fluorophore (~660 nm) in the region of FAP expression (actin filament, in red, on Figure 3a or membrane-tethered FAP on Figure 3b, yellow arrows). Treatment of T M R-para-MG stained cells that express intracellular membrane-displayed FAP with carbonyl cyanide m-chlorophenyl hydrazone (CCCP), a drug that causes rapid depolarization of mitochondria membrane potential[36], leads to rapid loss of TMR signal from the mitochondria (Figure 3b, SI Movie S2). Based on the spectroscopic properties of the dye, we attribute this loss of fluorescence to two phenomena: 1) loss of TMR-para-MG dye from the cell after depolarization of the mitochondria, as is seen for TMRM[37] (SI Figure S26) and 2) decrease of mitochondrial pH that leads to reversal of carbinol formation (SI Figure S21).
Another advantage of the TMR-para-MG dye is its usability for 2-photon imaging, which is known to be superior to confocal microscopy for imaging live and thick specimens. Because TMR has a good 2-photon cross-section[38], cells expressing FAPs and stained with TMR-para-MG are ~10-fold brighter under two photon excitation than the same cells stained with Cy3-MG or MG dye (Figure 3c).
In conclusion, we have developed a genetically targetable fluorescent probe TMR-para-MG that switches its fluorescence emission upon binding to a FAP, as well as a fluorogenic isomer TMR-meta-MG, which is efficiently activated by binding. These dyes penetrate through the plasma membrane effectively to stain different structures inside live cells. In cells that express FAP the TMR-para-MG dye labels target sites in one colour (MG emission) and mitochondria in another colour (TMR emission). Such dual colour emission of TMR-para-MG makes this dye a suitable tool for monitoring changes in mitochondrial membrane potential in response to drug treatment. In addition TMR-MG dyes have better 2-photon characteristics than MG dye alone or other tandem fluorophores that have previously been reported. Finally, the synthesis of multichromophore structures based on the TMR donor may result in cytosol directed genetically targeted light-harvesting materials that do not require cellular permeabilization for labeling. By applying approach described in this work it should be possible to synthesize colour-switching or fluorogenic tandem dyes by careful control of their geometry and interchromophore interactions.
Experimental Section
Details of the procedures for probes synthesis, including NMR, ESI, absorption and fluorescence characterization are provided in the Supporting Information. The procedures used for FAPs expression, cells transfection, mitochondria depolarization and fluorescence imaging are also described in details in the Supporting Information.
Supplementary Material
Acknowledgments
We thank Dr. Brigitte F. Schmidt for support in organic synthesis, Dr. Christopher Szent-Gyorgyi for assistance in yeast preparation and fruitful discussions, Dr. Gayathri C. Withers and Dr. Roberto R. Gil for the assistance in NMR experiments. NMR instrumentation at CMU was partially supported by NSF(CHE-0130903 and CHE-1039870). The multiphoton confocal microscope was purchased with support of the Gordon and Betty Moore Foundation. This work was supported in part by grants from the National Institutes of Health 7U54RR022241 and 1R01GM086237.
Footnotes
Supporting information for this article is available on the WWW under http://www.chembiochem.org or from the author
Contributor Information
Dr. Dmytro A Yushchenko, Email: ydmytry@gmail.com.
Prof. Dr. Marcel P Bruchez, Email: bruchez@cmu.edu.
References
- 1.Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Science. 1994;263:802. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 2.Shaner NC, Steinbach PA, Tsien RY. Nat Methods. 2005;2:905. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 3.Sample V, Newman RH, Zhang J. Chem Soc Rev. 2009;38:2852. doi: 10.1039/b913033k. [DOI] [PubMed] [Google Scholar]
- 4.Giepmans BN, Adams SR, Ellisman MH, Tsien RY. Science. 2006;312:217. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 5.Mehta S, Zhang J. Annu Rev Biochem. 2011;80:375. doi: 10.1146/annurev-biochem-060409-093259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rinkevich Y, Lindau P, Ueno H, Longaker MT, Weissman IL. Nature. 2011;476:409. doi: 10.1038/nature10346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao Y, Araki S, Wu J, Teramoto T, Chang YF, Nakano M, Abdelfattah AS, Fujiwara M, Ishihara T, Nagai T, Campbell RE. Science. 2011;333:1888. doi: 10.1126/science.1208592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brumbaugh J, Schleifenbaum A, Gasch A, Sattler M, Schultz C. J Am Chem Soc. 2006;128:24. doi: 10.1021/ja0562200. [DOI] [PubMed] [Google Scholar]
- 9.Diep CQ, Ma D, Deo RC, Holm TM, Naylor RW, Arora N, Wingert RA, Bollig F, Djordjevic G, Lichman B, Zhu H, Ikenaga T, Ono F, Englert C, Cowan CA, Hukriede NA, Handin RI, Davidson AJ. Nature. 2011;470:95. doi: 10.1038/nature09669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kralj JM, Hochbaum DR, Douglass AD, Cohen AE. Science. 2011;333:345. doi: 10.1126/science.1204763. [DOI] [PubMed] [Google Scholar]
- 11.Viviani D, Charlet A, van den Burg E, Robinet C, Hurni N, Abatis M, Magara F, Stoop R. Science. 2011;333:104. doi: 10.1126/science.1201043. [DOI] [PubMed] [Google Scholar]
- 12.Horie T, Shinki R, Ogura Y, Kusakabe TG, Satoh N, Sasakura Y. Nature. 2011;469:525. doi: 10.1038/nature09631. [DOI] [PubMed] [Google Scholar]
- 13.Larson DR, Zenklusen D, Wu B, Chao JA, Singer RH. Science. 2011;332:475. doi: 10.1126/science.1202142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yudushkin IA, Schleifenbaum A, Kinkhabwala A, Neel BG, Schultz C, Bastiaens PI. Science. 2007;315:115. doi: 10.1126/science.1134966. [DOI] [PubMed] [Google Scholar]
- 15.Karginov AV, Ding F, Kota P, Dokholyan NV, Hahn KM. Nat Biotechnol. 2010;28:743. doi: 10.1038/nbt.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lemke EA, Schultz C. Nat Chem Biol. 2011;7:480. doi: 10.1038/nchembio.620. [DOI] [PubMed] [Google Scholar]
- 17.Fernandez-Suarez M, Ting AY. Nat Rev Mol Cell Biol. 2008;9:929. doi: 10.1038/nrm2531. [DOI] [PubMed] [Google Scholar]
- 18.Patterson G, Davidson M, Manley S, Lippincott-Schwartz J. Annu Rev Phys Chem. 2010;61:345. doi: 10.1146/annurev.physchem.012809.103444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petchprayoon C, Yan YL, Mao S, Marriott G. Bioorg Med Chem. 2011;19:1030. doi: 10.1016/j.bmc.2010.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keppler A, Pick H, Arrivoli C, Vogel H, Johnsson K. Proc Natl Acad Sci U S A. 2004;101:9955. doi: 10.1073/pnas.0401923101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hein B, Willig KI, Wurm CA, Westphal V, Jakobs S, Hell SW. Biophys J. 2010;98:158. doi: 10.1016/j.bpj.2009.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnsson K, Maurel D, Banala S, Manley S. ACS Chem Biol. 2011 doi: 10.1021/cb2002889. [DOI] [PubMed] [Google Scholar]
- 23.Los GV, Encell LP, McDougall MG, Hartzell DD, Karassina N, Zimprich C, Wood MG, Learish R, Ohana RF, Urh M, Simpson D, Mendez J, Zimmerman K, Otto P, Vidugiris G, Zhu J, Darzins A, Klaubert DH, Bulleit RF, Wood KV. ACS Chem Biol. 2008;3:373. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- 24.Plass T, Milles S, Koehler C, Schultz C, Lemke EA. Angew Chem Int Ed Engl. 2011;50:3878. doi: 10.1002/anie.201008178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hinner MJ, Johnsson K. Curr Opin Biotechnol. 2010;21:766. doi: 10.1016/j.copbio.2010.09.011. [DOI] [PubMed] [Google Scholar]
- 26.Szent-Gyorgyi C, Schmidt BF, Creeger Y, Fisher GW, Zakel KL, Adler S, Fitzpatrick JA, Woolford CA, Yan Q, Vasilev KV, Berget PB, Bruchez MP, Jarvik JW, Waggoner A. Nat Biotechnol. 2008;26:235. doi: 10.1038/nbt1368. [DOI] [PubMed] [Google Scholar]
- 27.Fisher GW, Adler SA, Fuhrman MH, Waggoner AS, Bruchez MP, Jarvik JW. J Biomol Screen. 2010;15:703. doi: 10.1177/1087057110370892. [DOI] [PubMed] [Google Scholar]
- 28.Fitzpatrick JA, Yan Q, Sieber JJ, Dyba M, Schwarz U, Szent-Gyorgyi C, Woolford CA, Berget PB, Waggoner AS, Bruchez MP. Bioconjug Chem. 2009;20:1843. doi: 10.1021/bc900249e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szent-Gyorgyi C, Schmidt BF, Fitzpatrick JA, Bruchez MP. J Am Chem Soc. 2010;132:11103. doi: 10.1021/ja9099328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balzani V, Ceroni P, Maestri M, Vicinelli V. Curr Opin Chem Biol. 2003;7:657. doi: 10.1016/j.cbpa.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 31.Berera R, van Stokkum IH, Kodis G, Keirstead AE, Pillai S, Herrero C, Palacios RE, Vengris M, van Grondelle R, Gust D, Moore TA, Moore AL, Kennis JT. J Phys Chem B. 2007;111:6868. doi: 10.1021/jp071010q. [DOI] [PubMed] [Google Scholar]
- 32.Dichtel WR, Hecht S, Frechet JM. Org Lett. 2005;7:4451. doi: 10.1021/ol0516824. [DOI] [PubMed] [Google Scholar]
- 33.Rosch U, Yao S, Wortmann R, Wurthner F. Angew Chem Int Ed Engl. 2006;45:7026. doi: 10.1002/anie.200602286. [DOI] [PubMed] [Google Scholar]
- 34.Chazotte B. Cold Spring Harb Protoc. 2011;2011:895. doi: 10.1101/pdb.prot5641. [DOI] [PubMed] [Google Scholar]
- 35.Llopis J, McCaffery JM, Miyawaki A, Farquhar MG, Tsien RY. Proc Natl Acad Sci U S A. 1998;95:6803. doi: 10.1073/pnas.95.12.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim ML, Minamikawa T, Nagley P. FEBS Lett. 2001;503:69. doi: 10.1016/s0014-5793(01)02693-x. [DOI] [PubMed] [Google Scholar]
- 37.Ward MW. Methods Mol Biol. 2011;591:335. doi: 10.1007/978-1-60761-404-3_20. [DOI] [PubMed] [Google Scholar]
- 38.Albota MA, Xu C, Webb WW. Appl Opt. 1998;37:7352. doi: 10.1364/ao.37.007352. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.