Figure 4. A priori predictions of differentiated morphologies.
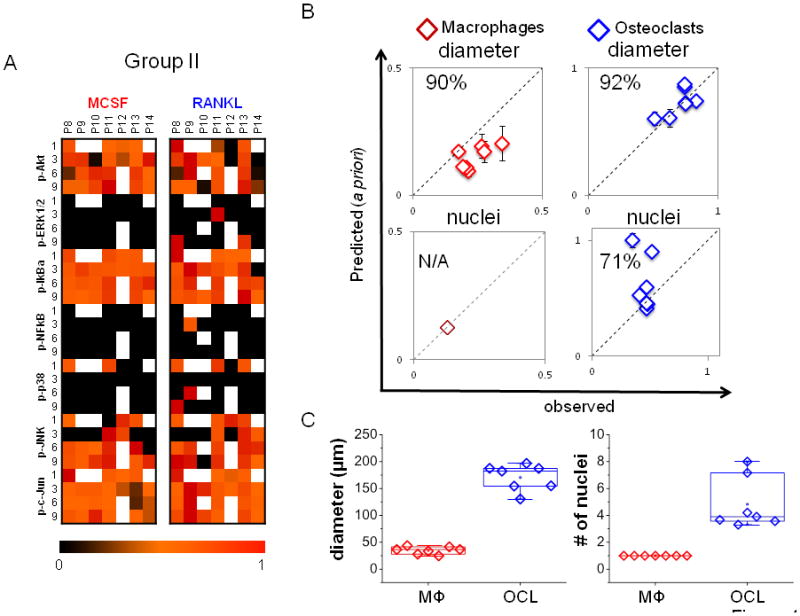
A) A compendium of time-dependent kinase signals of patients 8–14 (group II) used for a priori prediction is shown. B) The trained models for macrophages and osteoclasts generated with Group I data were used to predict cell diameter and number of nuclei from Group II kinase data. The model was effective in predicting cell diameter, with 90% predictability for macrophages and 91% predictability for osteoclasts, but only 71% for osteoclast nuclei. With only one nucleus, no variation can be predicted for macrophage. C) Experimental/observed quantification of mean diameter and average number of nuclei per patient (n=7).
