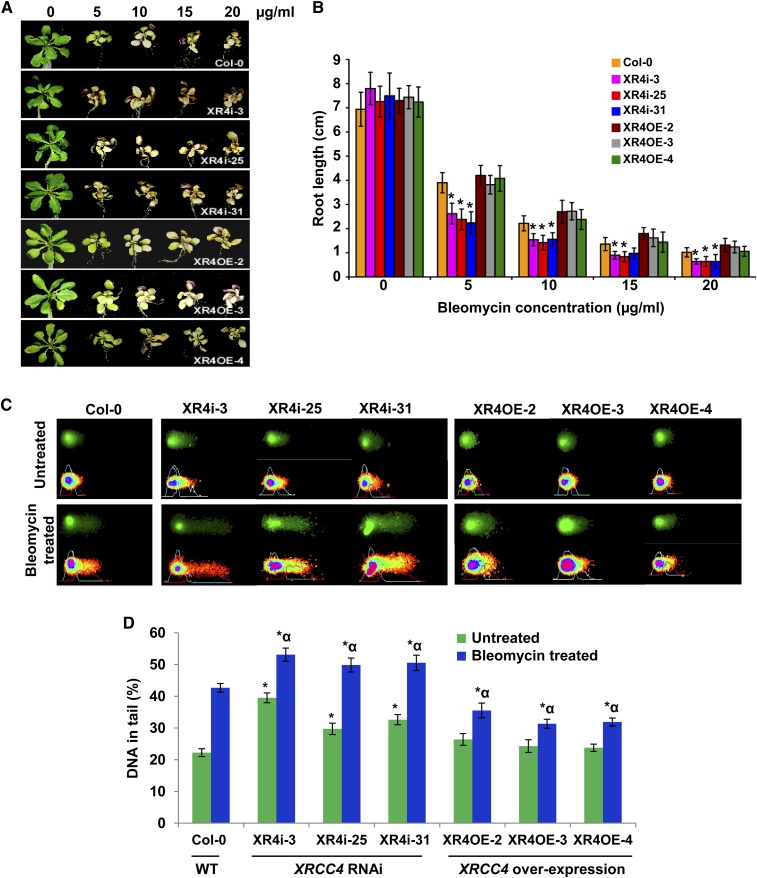
Figure 2.
Bleomycin Sensitivity and Comet Assays Show Hypersensitivity of XRCC4 RNAi Lines and Tolerance of XRCC4 Overexpression Lines to a DNA Damaging Chemical.
(A) Response of representative samples of Col-0, XRCC4 RNAi (XR4i), and overexpression (XR4OE) lines after exposure to bleomycin at noted concentrations. Photographs were taken at 10 d after exposure.
(B) Root length of above lines after exposure to bleomycin at various concentrations. Data are presented as mean ± sd (n = 5). Asterisks denote significant difference in the root length between XRCC4 RNAi lines versus Col-0 as determined by Student’s t test (P < 0.05).
(C) Microphotographs of nuclei in XRCC4 RNAi and overexpression lines by neutral comet assay with or without bleomycin (5 μg/mL) treatment. Nuclei with loss of round shape and smearing DNA fragments in the tail of the comet are considered damaged. The fraction of DNA in comet tails was visualized by SYBR Green staining (top panel), and quantitative assessment of DNA damage was performed using CometScore software (bottom panel)
(D) Quantification of the DNA fragments in tail (%) of 150 randomly selected cells per slide. Data are presented as mean ± sd. Asterisks indicate a significant difference between Col-0 and XRCC4 RNAi or overexpression lines without any bleomycin treatment, and asterisks with an alpha indicate a significant difference between Col-0 and XRCC4 RNAi or overexpression lines with 5 μg/mL bleomycin treatment according to Student’s t test (P < 0.05). WT, the wild type.