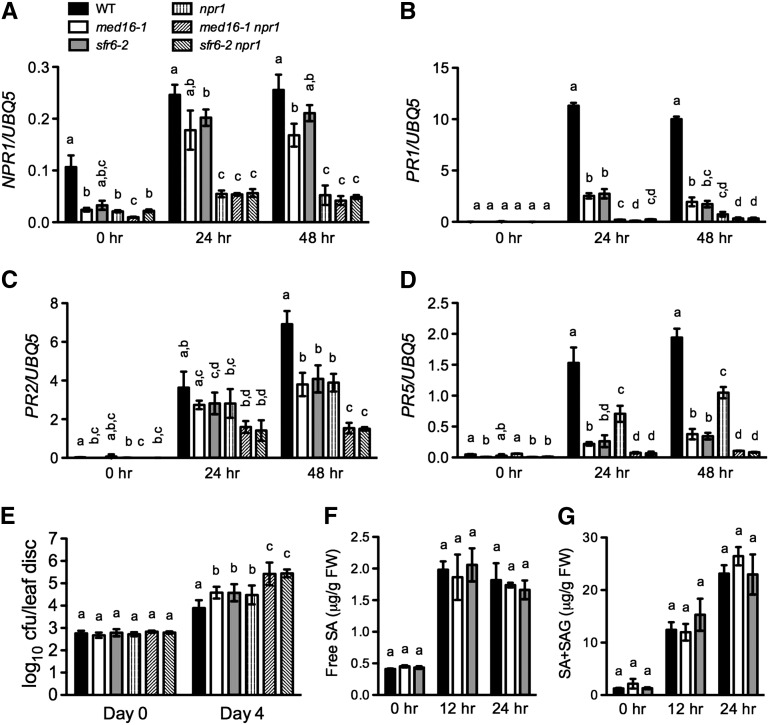
Figure 6.
Genetic Relationship between med16/sfr6 and npr1.
(A) to (D) Pst DC3000–induced expression of NPR1 (A), PR1 (B), PR2 (C), and PR5 (D) in med16/sfr6, npr1, med16/sfr6 npr1, and wild-type (WT) plants. Plants were inoculated with Pst DC3000 (OD600 = 0.001). Total RNA was extracted from the inoculated leaves collected at the indicated time points and analyzed for the expression of NPR1 using real-time qPCR. Expression was normalized against constitutively expressed UBQ5. Data represent the mean of three independent samples with sd.
(E) Growth of Pst DC3000 in med16/sfr6, npr1, med16/sfr6 npr1, and wild-type plants. Leaves of 4-week-old plants were inoculated with Pst DC3000 (OD600 = 0.0001). The in planta bacterial titers were determined immediately and 4 d after inoculation. Data represent the mean of eight independent samples with sd. cfu, colony-forming units.
(F) and (G) Free SA (F) and total (G) levels in Pst DC3000–infected med16/sfr6 and wild-type plants. Plants were inoculated with Pst DC3000 (OD600 = 0.001). The inoculated leaves were collected at the indicated time points for SA measurement. Data represent the mean of four independent samples with sd. Different letters above the bars indicate significant differences (P < 0.05, Student’s t test). The comparison was made separately among the wild type, med16-1, sfr6-2, npr1, med16-1 npr1, and sfr6-2 npr1 for each time point. All experiments were repeated with similar results. FW, fresh weight; SAG, SA-2-O-β-d-glucoside.