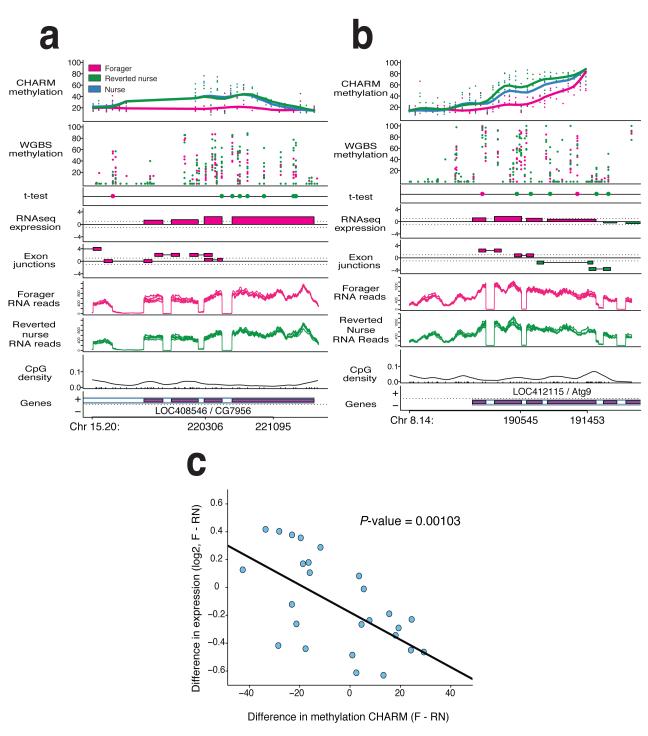
Figure 2. DNA methylation distinguishes nurses, foragers and reverted nurses.
Two examples of CHARM DMRs. a-b, Top panels show percent methylation for both CHARM and WGBS data sets, with points representing individual samples, and the smoothed lines representing the average for the phenotype. The t-test panel displays the top 1% differentially methylated CpGs by t-test. Color of the point indicates which phenotype has greater methylation at that CpG (n=6 per phenotype). The RNAseq expression panel is a t-statistic based on the number or reads detected within the annotated exons, with the color indicating the higher expressed phenotype. The Exon junctions panel is a t-statistic based on the number or reads detected spanning the exon junctions, as predicted by the TopHat program, with the color indicating the higher expressed phenotype. Switching between higher expressed nurse and forager exon junctions is indicative of alternative splicing events. The RNA reads panels indicate the number of reads per phenotype as compiled by TopHat program (n=6 per phenotype). The bottom two panels show the CpG density, and the relative position of the gene. c. Plot of relative gene expression comparing foragers to reverted nurses. 26 genes associated with DMRs were tested for gene expression differences by real-time PCR (n=12 per phenotype). The plot depicts the difference in average log2 expression versus average difference in methylation as determined by CHARM. Correlation analysis results in a P-value of 0.001.