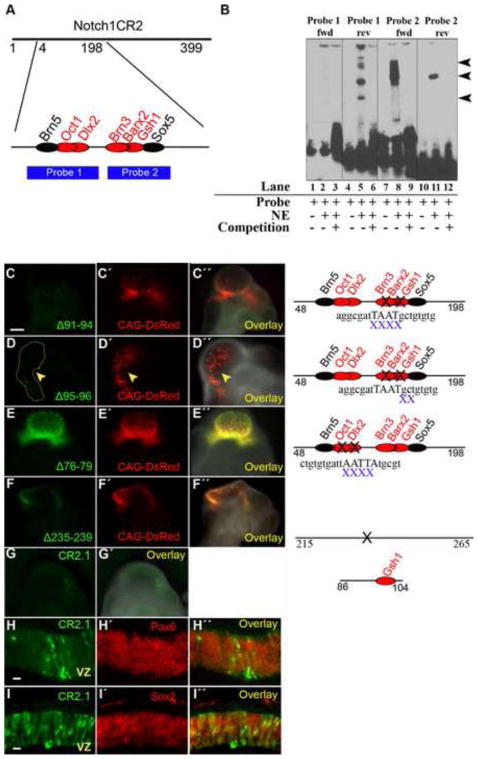
Figure 8. Gsh1 binding site in CR2 is important in directing GFP expression.
(A) Two probes (1 and 2) were designed between position 48 and 198 of CR2 which possessed TFBSs that are conserved between mouse and chick. TFBSs on forward and reverse strands are represented by red and black font, respectively. (B) Electrophoretic mobility shift assays (EMSA) show that DNA probes containing the Brn5/Oct1/Dlx2 (Probe 1) or Brn3/Barx2/Gsh1 (Probe 2) have CR2 sequence-specific binding activity with nuclear extract (NE) from E8 chick brain. The probes were run as forward strands (lanes 1–3, 7–9) and as reverse strands (lanes 4–6, 10–12). Lanes 1, 4, 7, and 10 are controls consisting of the probe alone without nuclear extract. Probe 1-rev, Probe 2-fwd, and Probe 3-rev showed band shift (lanes 5, 8, 11) that was competed away by the unlabeled competition probes (lanes 6, 9, 12). (C–F) Reporter assay using in ovo electroporation/transfection method with mutant CR2-GFP constructs generated by site-directed mutagenesis. (C) Deletion of position 91–94 (Δ91–94) removed the core binding sequence for the trans-acting factors of Brn3, Barx2, and Gsh1. The transfection of CR2Δ91–94 –GFP construct was not able to produce GFP expression in the E4 chick embryo. (D) Deletion of position 95–96 (Δ95–96) disturbed the peripheral binding sequence for Brn3, Barx2, and Gsh. The transfection of CR2-GFPΔ95–96 –GFP construct resulted in diminished GFP expression. (E) Deletion of position 76–79 removed the TFBS for Dlx2, which is a transcription factor of inhibitory neurons but is not conserved. The transfection of CR2-GFPΔ76–79 –GFP construct was able to drive GFP expression. (F) As a negative control, the deletion of position 235–239 (containing no known TFBS) on CR2 did not affect GFP expression. (G) Plasmid constructs containing subregions of CR2 were electroporated into the developing chick neural tube at E2, and GFP expression was detected at E4. CR2.1 (position 86–104) is an 18 bp fragment containing the conserved binding motif for Gsh1. (H–I) CR2.1 co-localizes with Pax6 and Sox2.