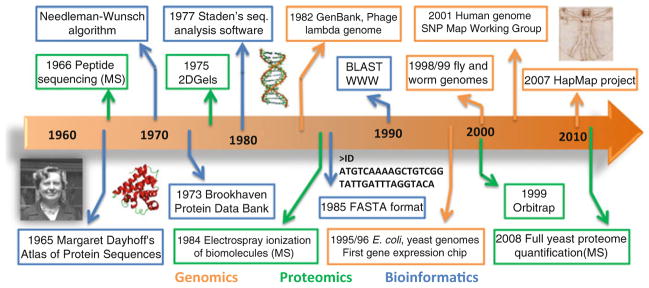
Fig. 7.1. Timeline of Bioinformatics, Genomic and Proteomic developments.
We selected and illustrated here several important landmarks in the development of genomics, proteomics, and bioinformatics, over the past 40 years. Examples of these include: the atlas of protein sequences, published as a book, by the bioinformatics pioneer Margaret Dayhoff; the first protein sequence analysis algorithms like the Needleman–Wunsch algorithm and analysis of gap-penalty costs by Haber and Koshland [87]; the creation of a protein structure repository (P.D.B-Protein Data Bank) that in 1974 contained atomic coordinates for 12 proteins; the first full genome sequences (1982–phage lambda, 1995–E. coli and 1996–budding yeast) along with the creation of the GenBank database that started with 606 sequences; the several technological developments in mass spectrometry (MS) like the first use of MS for peptide sequencing (1966), of electrospray ionization for biomolecules (1984), and novel ion traps like the orbitrap (1999) with increased mass accuracy and resolution, culminating in the first large-scale quantification of protein abundances of an eukaryotic cell (2008)