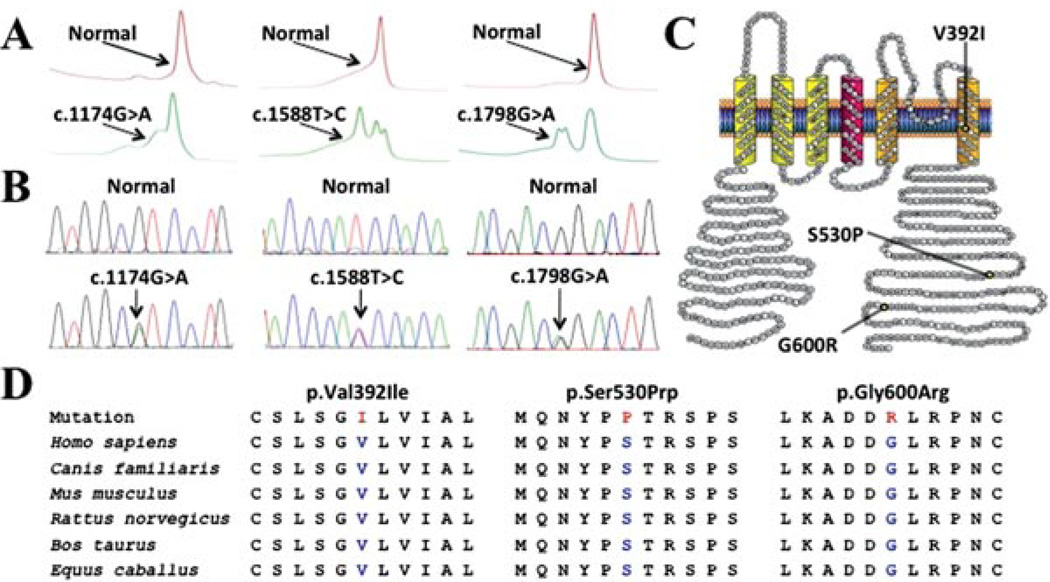
Figure 1.
Identification of SUDS- and SIDS-associated mutations in KCND3. Depicted are A: Denaturing high-performance liquid chromatography profiles (normal, red trace; and abnormal, green trace) and B: DNA sequencing chromatographs for c.1174G>A, c.1588T>C, and c.1798G>A nucleotide substitutions. C: Amino acid sequence conservation across species for p.Val392Ile, p.Ser530Prp, and p.Gly600Arg. D: Predicted protein topology schematic of Kv4.3, indicating the location of the p.Val392Ile, p.Ser530Pro, and p.Gly600Arg missense mutations.