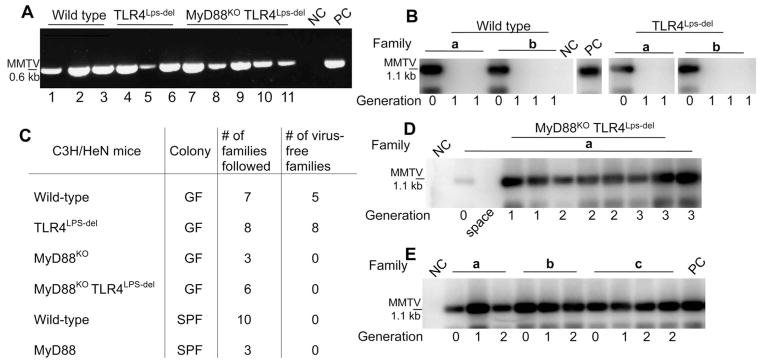
Fig. 2.
Differential MMTV persistence in GF and SPF mice of distinct genotypes. (A) Reverse transcription–PCR detection of MMTV virion RNA in the milk of GF C3H/HeN mice infected parenterally with MMTV(C3H). NC, negative control: RNA from the milk of an uninfected C3H/HeN mouse; PC, positive control: RNA from the milk of an SPF mouse injected with the same viral isolate. MMTV(C3H) and MMTV(LA) were used in two independent studies, data with MMTV(C3H) are shown. To ensure sufficient time for virus amplification in injected mice, the second litters by these dams were used for breeding and testing. (B) Splenic DNA from the first progeny (G1) of i.p. injected mice (G0) was subjected to MMTV-specific PCR, followed by Southern blot hybridization with an MMTV LTR–specific probe. Splenic DNA from an SPF G1 progeny of mice injected with the same virus isolate was used as PC, splenic DNA from an MMTV-negative C3H/HeN mouse was used as NC. a and b, independent families. The figure is assembled from nonconcurrent portions of the same image. (C) Virus fate in the offspring of parenterally infected (G0) mice of different genotypes and maintenance conditions. Summary of data from MMTV-infected mice, showing virus loss or persistence at G1. (D) Transmission of infectious MMTV(C3H) in GF MyD88KOTLR4Lps-del mice. Detection of integrated proviruses was done as in (B). NC, DNA from the spleen of an uninfected C3H/HeN mouse. A single representative family (a) is shown. (E) Transmission of infectious MMTV(C3H) in ASF-associated gnotobiotic wild-type C3H/HeN mice as detected by MMTV(C3H)-specific PCR, followed by Southern blot analysis, performed with splenic DNA of mice from G0 to G2. a to c, different families. Splenic DNA from SPF G1 progeny of mice injected with the same virus isolate served as PC, and splenic DNA from an MMTV-negative C3H/HeN mouse was used as NC. For (A), (B), (D), and (E), PCRs were performed under saturating conditions.