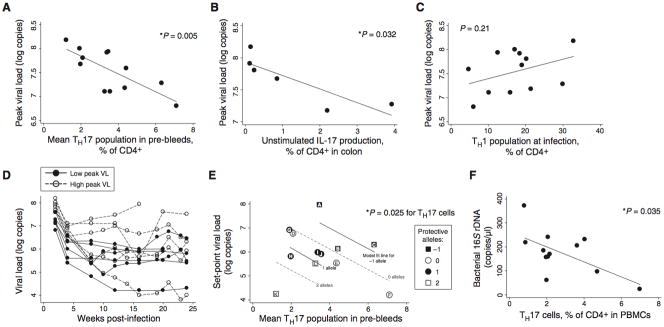
Fig. 2.
Macaques with large pre-existing Th17 compartments develop lower viral loads. (A) The Th17 population measured in pre-bleeds is predictive of peak viral loads, reached two weeks after infection. The relationship remains significant when any single animal is eliminated from the analysis. (B) Production of IL-17 by unstimulated, colon-resident lymphocytes is also predictive of peak viral loads. Biopsies were taken before infection in six animals that were followed in Figure 1 (see Figure 1G). (C) The Th1 population (CD3+CD4+IFN-γ+IL-17−) present before infection was not predictive of viral loads. (D) Animals with a high peak viral load (peak viral load above the median; shown with open circles and dashed lines) did not always develop a high set-point; animals with a low peak viral load (below the median; closed circles and solid lines) did not always develop a low set point. (E) Th17 cells and MHC alleles were jointly predictive of set-point viral loads. Multiple regression of set-point viral loads against Th17 cells and protective MHC alleles (count of alleles; see Table S1) demonstrated significant predictive value overall (p=0.023 for multivariate F-test) and for each variable (p=0.025 and 0.009 for Th17 cells and MHC alleles, respectively). Letter labels provide a cross reference to MHC allele information detailed in Table S1. (F) The percentage of Th17 cells among CD4+ T cells is a significant predictor of bacterial 16S rDNA at 2 weeks after infection.