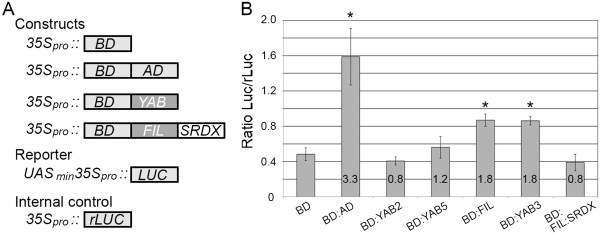
Figure 2.
Transcriptional activities of vegetatively expressed YAB proteins. (A) Outline of constructs used for transactivation assays. AD, GAL4 activation domain; BD, GAL4-DNA binding domain; YAB, vegetatively expressed YABs (FIL, YAB2, YAB3, YAB5); SRDX, repressive domain (see text for details); UAS, BD binding site; LUC, Firefly Luciferase; rLUC, Renilla Luciferase. (B) YAB transcriptional activation assays with Gal4 BD and BD-AD used as negative and positive control respectively. rLuc was used as an internal control to determine the relative bioluminescence for each sample (ratio Luc/rLuc). Numbers in the shaded boxes indicate fold activation as calculated by dividing total Luc activity of samples by the baseline values arising from the 35Spro::BD construct. Asterisks indicate significant differences (Student's t-test; p < 0.05) and error bars indicate SEM.