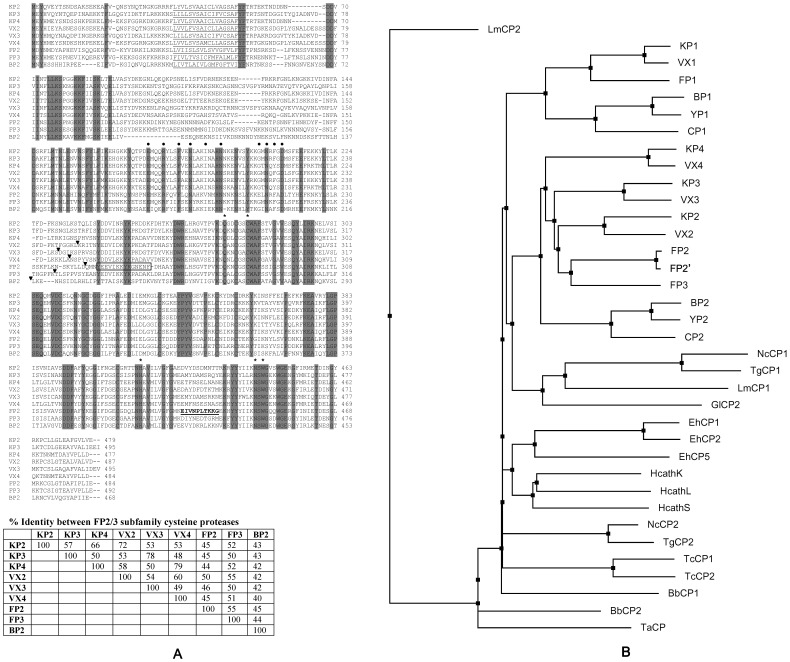
Figure 1. Sequence alignment of knowpains. A.
The deduced amino acid sequences of knowpain-2 (KP2), knowpain-3 (KP3), knowpain-4 (KP4), vivapain-2 (VX2), vivapain-3 (VX3), vivapain-4 (VX4), falcipain-2 (FP2), falcipain-3 (FP3) and bergheipain (BP2) were aligned by using the multalin program. Predicted transmembrane domains are underlined, and ERFNIN and GNFD inhibitory motifs in the prodomain are labeled with filled circles. The positions of mature domain processing sites are indicated by arrowheads, and the catalytic amino acids are indicated by asterisks. The refolding domain and haemoglobin-binding motif of FP2 are boxed and underlined, respectively. The table shows % sequence identities within the FP2/3 subfamily based on the entire prodomain-mature protease regions. B. The phylogram was generated using the Neighbor Joining algorithm from the sequence alignment of mature protease domains (beginning with the conserved DWR motif to the end) of papain-like proteases of parasitic Protozoa and their selected human homologs (cathepsin L, cathepsin K, and cathepsin S). The accession numbers for various proteases are: P. knowlesi proteases knowpain-1 (KP1, XP_002260291), knowpain-2 (KP2, XP_002259153), knowpain-3 (KP3, XP_002259152), knowpain-4 (KP4, XP_002259151); P. falciparum proteases falcipain-1 (FP1, XP_001348727, falcipain-2 (FP2A XP_001347836), falcipain-2′ (FP2′, XP_001347832), falcipain-3 (FP3, XP_001347833); P. vivax proteases vivapain-1 (VX1, XP_001615807), vivapain-2 (VX2, XP_001615274), vivapain-3 (VX3, XP_001615273), vivapain-4 (VX4, XP_001615272); P. berghei proteases bergheipain-1 (BP1 XP_677643) and bergheipain-2 (BP2, PBANKA_093240); P. chabaudi chabaudi proteases chaubipain-1 (CP1, AAP43629) and chaubipain-2 (CP2, AAP43630); P. yoelii yoelii proteases yoelipain-1 (YP1, XP_729023) and yoelipain-2 (YP2, XP_726900); Babesia bovis proteases bovipain-1 (BV1, XP_001612131) and bovipain-2 (BV2, XP_001610695); Entamoeba histolytica cysteine protease-1 (EhCP1, Q01957), cysteine protease-2 (EhCP2, Q01958), cysteine protease-5 (EhCP5, CAA62835); Giardia lamblia cysteine protease-2 (GlCP2, EAA41050); Leishmania major cysteine protease-1 (LmCP1, AAB48119), cysteine protease-2 (LmCP2, AAB48120); Neospora caninum cysteine protease-1 (NcCP1, CCA30060) cysteine protease-2 (NcCP2, CBZ49954); Theileria annulata cysteine protease (TaCP, AAA30135); Toxoplasma gondii cysteine protease-1 (TgCP1, AAL60053), cysteine protease-2 (TgCP2, ABY58967); Trypanosoma cruzi cysteine protease-1 (TcCP1, AAA30181), cysteine protease-2 (TcCP2, AAC37213); human cathepsin L (HcathL), AAC23598), cathepsin S (HcathS, AAC37592) and cathepsin K (HcathK, P43235).