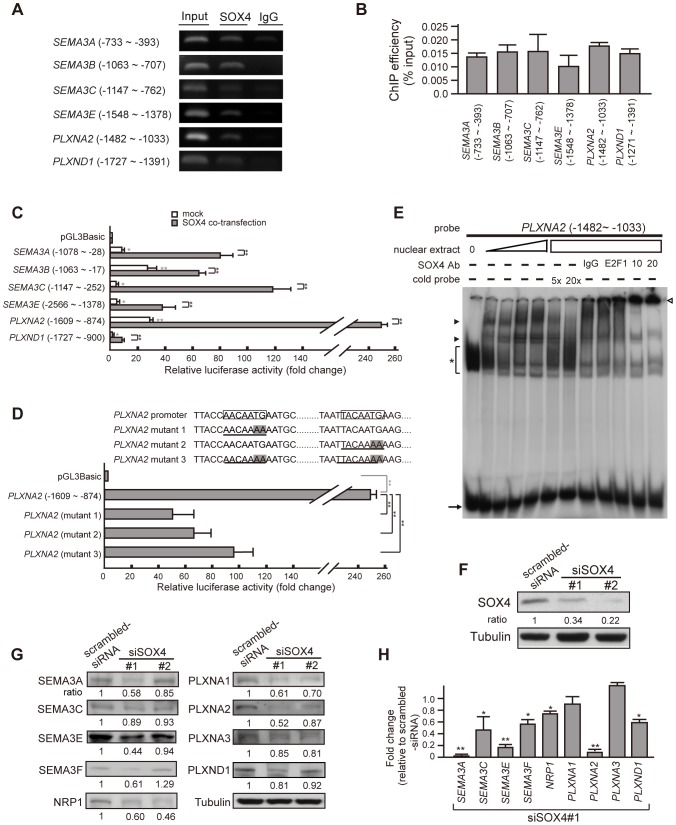
Figure 3. SOX4 trans-activates the transcription of SEMA3, Plexin, and NRP1.
(A) representative end-point PCR of chromatin immunoprecipitation (ChIP) using an antibody against SOX4 in PANC-1 cells. Genomic DNAs were amplified with primer sets after bound proteins were digested with proteinase K. The ChIP results showed binding of SOX4 to the genomic region harboring SOX4 binding sites in the predicted promoter of most SEMA3 and Plexin genes. (B) quantification of ChIP results in (A), n> = 3. Each lane in figure 3A was given a value measured by densitometry and normalized with the value of IgG. ChIP efficiency is defined as percentage of the value derived from PCR of SOX4-immunoprecipitate relative to that of total genomic input. Error bars, standard deviation (SD) from at least triplicate. (C) increased luciferase activity of constructs containing SEMA3- or Plexin- promoter in the absence (white bars) or presence (gray bars) of co-expressed SOX4 in comparison with the promoterless pGL3-Basic control. The constructs harboring a SOX4-consensus binding site were transiently transfected into SOX4-containing HeLa cells. Shown are ratios of firefly luciferase expression to Renilla luciferase expression (expressed from co-transfected plasmid), measured 48 h after transfection, normalized to the mean ratio from the pGL3-Basic. Error bars show SD, n> = 4. Student's t-test, *, P<0.01; **, P<0.001. In the presence of co-transfected SOX4, luciferase activity driven by each SEMA3- or Plexin- promoter is further augmented (gray bars) compared to mock-transfected (white bars) and controls. (D) Transcriptional activity of PLXNA2 promoter driven by SOX4 is attenuated when the consensus SOX4-binding sites are mutated. Boxed DNA sequence, the consensus SOX4-binding site; Underline with shadow in letters, the mutated SOX4-binding sites. Bar graph shows that mutation of either SOX4-binding site caused significant decrease in luciferase activity driven by PLXNA2 promoter in the presence of overexpressed SOX4. Error bars, SD, n = 5. Student's t-test, *, P<0.01; **, P<0.001. (E) EMSA shows that the migration of a radiolabeled DNA fragment derived from the PLXNA2 promoter is retarded by SOX4-containing HeLa crude nuclear extract in a concentration-dependent manner. The signal of protein-bound retarded band (arrowheads) was specifically diminished by cold probes containing SOX4-binding consensus sequences and was super-shifted by anti-SOX4 antibody (empty arrowhead). Asterisk, non-specific binding signals; arrow, un-bound radiolabeled probe. (F) effective RNAi knockdown of endogenous SOX4 in PANC-1 cells shown by SOX4 immunoblotting. The value indicated is the normalized relative intensity measured by densitometry (The ratio SOX4/Tubulin for scrambled-siRNA cells is defined as 1.). Two stable siSOX4 clones labeled as siSOX4#1 and siSOX4#2 were constructed with the targeting oligonucleotide directed against different nucleotide sequences in SOX4 gene (siSOX4#1, nt.1999–2019 NM_003107.2; siSOX4#2, nt.1362–1382 NM_003107.2). (G) Immunoblotting in SOX4-depleted cells shows a concomitant decrease in the protein amount of NRP1, SEMA3- and Plexin- family members. Tubulin was used as a standard for normalization in the quantification of each lane measured by densitometry. The ratio indicated was the normalized value in siSOX4 cells relative to the normalized value in scrambled-siRNA cells. (H) Real-time PCR to quantify mRNA expression of SEMA3 and PLXN genes in SOX4-depleted cells relative to SOX4-abundant scrambled-siRNA cells (shown by fold-change). Student's t-test, *, P<0.01; **, P<0.001.