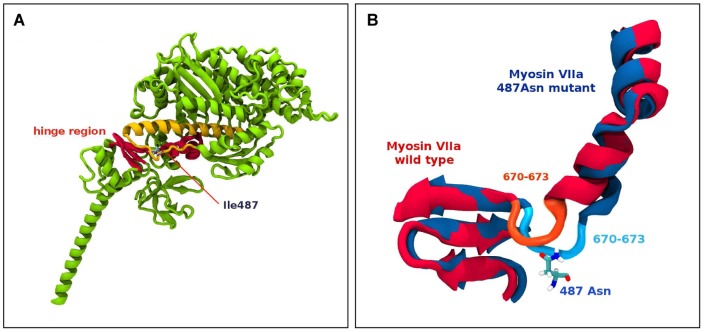
Figure 3. Molecular modelling of the Myo7aI487N/I487N ewaso mutation.
(A) Partial ribbon representation of a 3D model of myosin V 2DFS highlighting the Ile487 amino acid residue, which is localised to the hinge region between the head and tail domains of the protein. (B) Schematic diagram showing superimposed images of the hinge region (Myo7a+/+ in red and the Myo7aI487N/I487N ewaso mutation in blue) after 4 ns of molecular dynamics simulation showing a conformational change in the hinge region close to the Ile487 mutation site, showing the involvement of residues 670–673 around the hinge region causing the distortion.