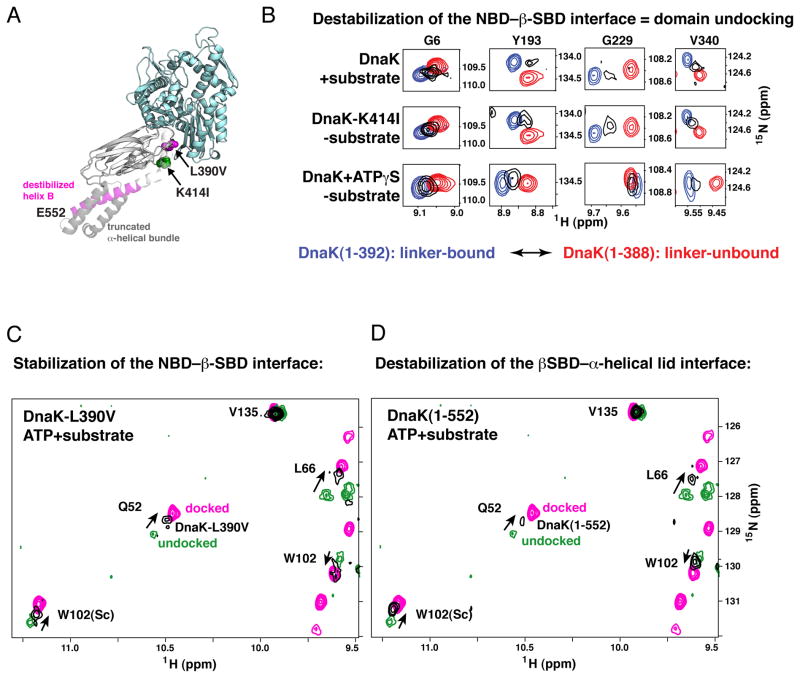
Figure 5. The Impact of Competition Between the NBD–β-SBD and β-SBD–α-Helical Lid Interfaces on the Hsp70 Allosteric Landscape.
(A) DnaK sequence modifications that result in perturbations in its conformational ensemble are mapped onto the modeled structure of the allosterically active conformation (modeled as for Figure 4): L390V and C-terminal truncations, which favor domain docking, are shown in magenta, K414I, which favors domain undocking, is shown in green.
(B) Destabilization of the NBD–β-SBD interface results in domain undocking even in the absence of substrate: Blow-up of the amide-TROSY spectra of DnaK(1-392) (blue, representing the NBD linker-bound state) and DnaK(1-388) (red, representing the NBD linker-unbound state) overlaid on the spectra of two-domain DnaK under conditions that stabilize domain undocking, either upon substrate binding to the ATP-bound DnaK(1-605) (top panels) or upon perturbation of the NBD–β-SBD interface, viz. ATP-bound DnaK(1-552)K414I (middle panels), or ATPγS-bound DnaK(1-552) (bottom panels). Resonances shown (Gly6, Tyr193, Gly229, and V340) report on long-range conformational changes in the nucleotide-binding site upon linker binding to the NBD (see Supplemental Experimental Procedures). The spectra of the isolated NBD constructs are with the corresponding nucleotide bound (ATP, top and middle panels) or ATPγS-bound (bottom panels).
(C) Stabilization of the NBD–β-SBD interface in DnaK(1-605)L390V or (D) destabilization of the β-SBD–α-helical lid interaction in DnaK(1-552) favors the domain-docked conformation even in the presence of substrate. A representative region of the amide-TROSY spectra of the ATP-bound state of DnaK(1-552) (magenta; the domain-docked conformation) and ATP-/substrate-bound DnaK(1-605) (green; the domain-undocked ensemble of linker-bound and linker-unbound conformations) overlaid with spectra of DnaK(1-605)L390V (C) and DnaK(1-552) (D), shown in black. Consistent with previous biochemical studies (Kumar et al., 2011; Swain et al., 2007) neither the L390V mutation on the NBD–β-SBD interface nor disruption of the β-SBD–α-helical lid interface in DnaK(1-552) affects the ATP-bound conformation in the absence of substrate.