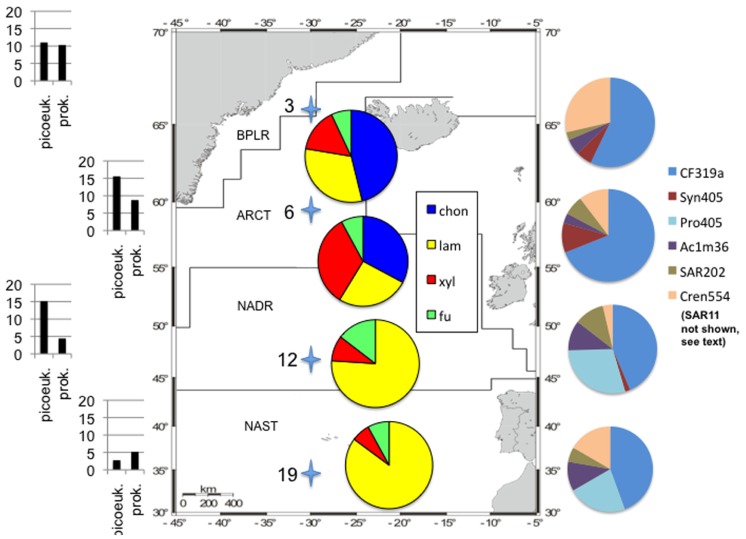
FIGURE 2.
Study area with oceanic provinces, sampling stations, enzymatic hydrolysis patterns, cell counts, and community composition information. Station locations shown as stars (station numbers on left); map modified from Gomez-Pereira et al. (2010). Cell counts on left side of figure: total prokaryotic cell counts as “prok,” picoeukaryotes counted via flow cytometry as “picoeuk.” Left hand scale: for picoeukaryotes, ×103 cells ml−1, for prokaryotes, ×105 cell ml−1. Data from Schattenhofer et al. (2011) and Gomez-Pereira et al. (2010). Pie charts superimposed on map: relative contribution of each enzyme activity to total activity measured at a station; substrate names as in Figure 1. Pie charts on right hand side: relative contribution of FISH-stained cells to total cell counts from water samples that were not subject to flow cytometry (“unsorted cells”); data from Schattenhofer et al. (2011) SI Table 2. Note that SAR11 cells contributed 25–32% of total DAPI-stained cells at each station, and are not plotted here for clarity. Pie charts as shown represent 17.5, 29, 27.5, and 18% of the total DAPI-stainable cells at S3, S6, S9, and S19, respectively. CF319a, Bacteroidetes; SYN405, Synechococcus; PRO405, Prochlorococcus; AC1M36, marine clade I Actinobacteria; SAR202, SAR 202 clade; CREN554, Crenarchaeota marine group I.