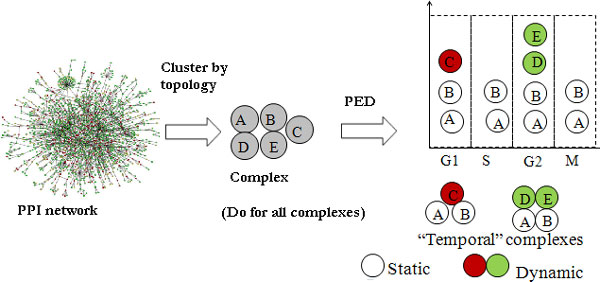
Figure 2.
Workflow for the incorporation of time information into complexes. We first cluster the PPI network using existing topology-based methods; here we used MCL-CAw [20]. For the clusters that do not match or just partially match known complexes in the Wodak catalogue [21], we incorporate temporal information (in the form of cell cycle phase labels) to check whether the cluster is a fusion of multiple time-based complexes. For this we bin the constituent proteins into the four phases (G1, S, G2, M) based on their phase labels. This procedure helps to decompose large clusters into distinct time-based complexes.