Abstract
Background
MicroRNAs (miRNAs) have been known to play an important role in several biological processes in both animals and plants. Although several tools for miRNA and target identification are available, the number of tools tailored towards plants is limited, and those that are available have specific functionality, lack graphical user interfaces, and restrict the number of input sequences. Large-scale computational identifications of miRNAs and/or targets of several plants have been also reported. Their methods, however, are only described as flow diagrams, which require programming skills and the understanding of input and output of the connected programs to reproduce.
Results
To overcome these limitations and programming complexities, we proposed C-mii as a ready-made software package for both plant miRNA and target identification. C-mii was designed and implemented based on established computational steps and criteria derived from previous literature with the following distinguishing features. First, software is easy to install with all-in-one programs and packaged databases. Second, it comes with graphical user interfaces (GUIs) for ease of use. Users can identify plant miRNAs and targets via step-by-step execution, explore the detailed results from each step, filter the results according to proposed constraints in plant miRNA and target biogenesis, and export sequences and structures of interest. Third, it supplies bird's eye views of the identification results with infographics and grouping information. Fourth, in terms of functionality, it extends the standard computational steps of miRNA target identification with miRNA-target folding and GO annotation. Fifth, it provides helper functions for the update of pre-installed databases and automatic recovery. Finally, it supports multi-project and multi-thread management.
Conclusions
C-mii constitutes the first complete software package with graphical user interfaces enabling computational identification of both plant miRNA genes and miRNA targets. With the provided functionalities, it can help accelerate the study of plant miRNAs and targets, especially for small and medium plant molecular labs without bioinformaticians. C-mii is freely available at http://www.biotec.or.th/isl/c-mii for both Windows and Ubuntu Linux platforms.
Background
MicroRNAs (miRNAs) are a class of small, non-coding, single-stranded RNA molecules of 18-22 nucleotides. In various species, they play roles in gene regulation by targeting mRNAs at the post-transcriptional level [1]. In plants, miRNAs are involved in organ development and environmental responses [2-4]. Although several miRNA and target prediction tools are available [5,6], the number of tools customized for plant miRNA and target analysis is limited. Among them are microHARVESTER, a web server for identifying plant miRNAs [7]; the miRU [8], psRNATarget [9], and TAPIR [10] web servers for identifying plant miRNA targets; a web-based toolkit for the analysis of plant small RNAs [11]; and the miRTour web server for plant miRNA and target prediction [12]. Most of these public web servers limit the number of input sequences and focus on only miRNA or target identification. Target-align [13] was recently proposed for plant miRNA target identification and developed as both web and command line versions. Even though several studies in computational identification of plant miRNAs and their targets are available [14-25], their methods were mainly presented as flow diagrams of connected programs (e.g., BLAST [26], UNAFold [27]). To follow the same steps, users need to install these programs, understand their usage, comprehend the meaning and format of the results, and have the programming experience for connecting them together.
Taking into account all these computational steps and criteria for plant miRNA and target identification [28-38], we developed C-mii, a standalone software package with graphical user interfaces for identifying, manipulating, and analyzing plant miRNAs and targets. C-mii is implemented as an all-in-one Java package weaving together sequence similarity search, secondary structure folding, automatic stem-loop identification and manipulation, and functional and gene ontology (GO) annotation. In addition, it comes pre-installed with databases of proteins, non-coding RNAs, and mature miRNAs. C-mii expects a set of nucleotide sequences (e.g., cDNAs, Expressed Sequence Tags (ESTs), Genome Survey Sequences (GSS)) in FASTA format as input. The identification steps are divided into miRNA and target identification pipelines. Users can customize parameter settings for each step of the identification, and filter and manipulate the results according to various biological criteria.
Materials and methods
Workflow overview
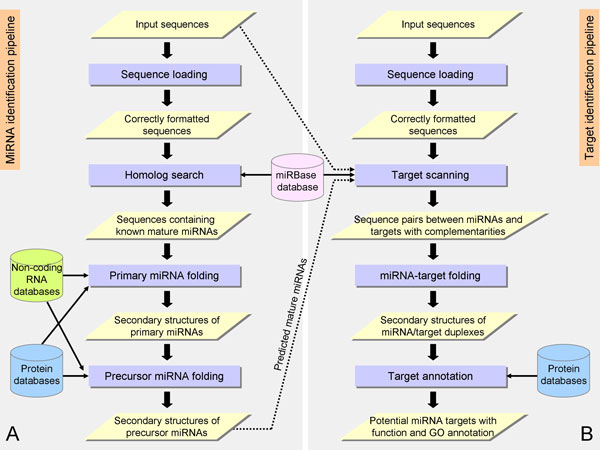
Figure 1 shows the overall computational steps of C-mii with two pipelines consisting of miRNA and target identifications. The miRNA identification pipeline includes the consecutive execution of four main modules: sequence loading and validation, homolog search, primary miRNA folding, and precursor miRNA folding. The sequence validation checks the uploaded file format and excludes sequences longer than 3000 nt or with non-nucleotide characters. The homolog search determines whether input sequences contain homologous mature miRNAs. This module is implemented based on BLAST and sequence scan with user-selected mature miRNAs pre-installed from miRBase [39,40]. The primary miRNA folding module predicts the secondary structure folding of input sequences containing homologous miRNAs. Precursor miRNA folding, the last module of the pipeline, extracts and re-predicts the stem-loop structures from the primary miRNA structures. Then, for validation, it examines whether these structures satisfy the constraints of plant precursor miRNA biogenesis. Both the primary and precursor miRNA folding modules employ UNAFold to predict secondary structure folding. The Rfam [41] and UniProt [42] databases are also incorporated, with each step allowing users to remove sequences that are other types of RNAs and protein-coding sequences, respectively.
Figure 1.
Workflow of C-mii. (A) miRNA identification pipeline, and (B) target identification pipeline. The dashed lines represent the optional usage of input sequences and predicted mature miRNA sequences from the miRNA identification by the target scanning.
The target identification pipeline consists of four consecutive modules: sequence loading and validation, target scanning, miRNA-target folding, and target annotation. The sequence loading module is the same as that for miRNA identification, but accepts input sequences of longer lengths (≤ 20,000 nt). The target scanning module determines if input sequences contain the complementary sites to mature miRNAs of interest. Implementation of this module is similar to the homolog search except for the use of reverse-complement mature miRNAs as queries for BLAST. The miRNA-target folding module utilizes UNAFold to predict the secondary structure and free energy of miRNA:target duplexes. This module was introduced to C-mii to help refine the target identification result. The last module, target annotation, supplies function and gene ontology (GO) for the potential target sequences. Along the computational steps of both pipelines, users can customize parameters for their execution, select databases of interest, and explore and filter the results.
Pre-installed databases
The pre-installed databases of C-mii are divided into three categories. First, the mature miRNA database, miRBase release 16, was incorporated for homolog search. Second, the non-coding RNA database, Rfam 10 with removed miRNAs, was introduced for removing other types of RNAs. Third, the protein databases, the UniProtKB/Swiss-Prot release 2010_12 and UniProtKB/TrEMBL release 2011_01, were incorporated for removing protein-coding sequences in the primary and precursor miRNA folding steps and for identifying gene functions during target annotation. These databases were pre-processed using the formatdb program in the BLAST package. Users can update these databases from our web site via a menu in C-mii. Furthermore, users can integrate their own protein and non-coding RNA databases into the system by following the pre-processing steps documented on the C-mii web site.
Pre-installed software packages
To ease its installation and usage, C-mii was designed as a complete package containing all required software, including BLAST, Java Development Kit (JDK), Perl, Python, UNAFold, and Ghostscript [43], which can be customized during installation. To explore conservation and co-evolution among the predicted and known precursors or mature miRNAs of an miRNA family, CLUSTAL W [44], MUSCLE [45] and Jalview [46] were pre-installed for performing and visualizing the multiple sequence alignment of selected identification results. In addition, we have deployed prefuse visualization toolkit [47], ICEpdf Viewer [48], and JFreeChart [49] for visualizing GO trees, the secondary structure folding of primary and precursor miRNAs, and the infographics, respectively.
Results and discussion
C-mii is composed of two pipelines for plant miRNA and target identifications, which could be used autonomously or consecutively. The functionalities of these pipelines have been described with biological rationales and a running example (visit http://www.biotec.or.th/isl/c-mii/documentation.php under "C-mii running example section" to see screenshots of all steps).
MiRNA identification pipeline
Taking into account the computational steps and criteria for plant miRNA identification as described previously [28,29,31], the miRNA prediction menu consists of four consecutive submenus starting from sequence loading, homolog search, primary miRNA folding, and precursor miRNA folding. Users need to build a new project before uploading nucleotide sequences in a FASTA file. Sequences longer than 3,000 nt or containing characters other than A, T, C, G, U, and N will be excluded. As a running example, we built a TAIR10 cDNA project of 33,602 sequences. Due to sequence validation, 30,707 sequences remained as input for the homolog search.
Homolog search
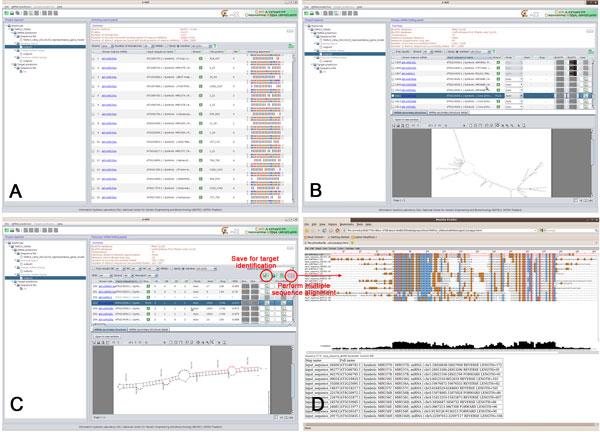
The homolog search module helps users identify input sequences that contain mature miRNA sequences from miRBase. In this step, users can select mature miRNAs of multiple plants from miRBase to be used as source mature miRNAs for the identification process. The identification methods include sequence scans with and without BLAST. The sequence scan with BLAST is much faster, but with the trade-off of possibly missing matches due to the word size limitation of BLASTN, which needs to be 4 or greater. Users can also customize the E-value (≤ 10 by default) of BLASTN, the number of allowed mismatches (≤ 4 by default) between a source mature miRNA and its homolog in an input sequence, and the number of processors automatically detected by C-mii for running the homolog search. Figure 2A shows the homolog search results for Arabidopsis mature miRNAs from miRBase with the 30,707 TAIR10 cDNA sequences with default parameter settings. Using the plus strand only, 1286 Arabidopsis cDNAs of 129 miRNA families and 231 members were identified. We selected all these sequences as input for the primary miRNA folding module.
Figure 2.
MiRNA identification results. (A) homolog search results, (B) primary miRNA folding results, (C) precursor miRNA folding results with various filters corresponding to suggested criteria in previous literatures, (D) multiple sequence alignment of known and predicted precursor miRNAs of miR156 family.
Primary miRNA folding
An miRNA gene needs to be non-coding and have a stem-loop precursor in its secondary structure [31]. The primary miRNA folding module helps users remove protein-coding sequences and other types of non-coding RNAs from input sequences, and predicts the secondary structure folding of primary miRNAs (pri-miRNAs) from the remaining sequences. The removal of unwanted sequence types is based on BLASTX and BLASTN against protein and non-coding RNA databases, selectable by users. Users can also adjust the E-value to limit the number of search results. The lower the E-value, the larger the number of sequences remains for secondary structure folding. C-mii sets the default E-value of BLASTN against Rfam as 1E-8 according to our previous benchmark [50]. Users may also customize the folding temperature, maximum base pair distance, and maximum bulge or interior loop size of UNAFold.
Figure 2B shows the primary miRNA folding results with default parameter settings with the exception of the BLASTX E-value ≤ 1E-5. Sequences with a clickable BLASTN or BLASTX box are sequences that hit with other sequence types. By clicking on these boxes, users can explore their E-values and hit sequences. The "Only results" check box allows users to filter out these sequences. For the remaining sequences, users can interactively explore their secondary structures and minimal free energies (MFEs) predicted by UNAFold, minimal folding free energy indices (MFEI) [30], sequence lengths, and nucleotide and GC content. The "Mode" column provides two options for extracting stem-loop precursors from a secondary structure of a primary miRNA. C-mii decides the cleavage positions for users in Auto mode. The Manual mode allows users to specify the start and stop cleavage positions. From the results of our running example using the plus strand only, 567 Arabidopsis cDNAs in 124 miRNA families and 224 members remained. All these sequences were selected with Auto mode for the precursor miRNA folding module.
Precursor miRNA folding
The precursor miRNA folding module helps users (1) extract the stem-loop structures from the secondary structures of pri-miRNAs, (2) remove stem-loop sequences that hit protein-coding sequences and other types of non-coding RNAs, (3) re-predict the secondary structure folding of the extracted stem-loop sequences, and (4) verify the predicted structures with previously suggested criteria. With the Auto mode setting from the previous step, the precursor miRNA folding module will cleave a pri-miRNA structure from the start position of the found homologous miRNA to the end position of its duplex miRNA* with two-nucleotide 3' overhangs. The extracted stem-loop sequences are screened against the protein and non-coding RNA databases again. UNAFold is then reapplied to the remaining sequences to predict the secondary structure folding of precursor miRNAs (pre-miRNAs). Users can customize the same set of parameters as in the primary miRNA folding step.
Figure 2C shows the precursor miRNA folding results with default parameter settings with the exception of the BLASTX E-value ≤ 1E-5. Based on previously reported criteria [28,31], structures with multi-loops or mature miRNA sequences not located within one arm will be automatically removed by C-mii. Besides MFEIs, users can filter the results by restricting the number of two-nucleotide 3' overhangs, the number of mismatches between miRNA:miRNA* duplexes, the number of bulges, and bulge sizes as proposed in [31]. Users can browse through the predicted secondary structures, select potential miRNAs based on filters, save them for target identification, and export them as an archive. Users may also perform multiple sequence alignments among the identified and known precursor or mature miRNAs of the same family to explore their conservation and evolution (Figure 2D). Using the plus strand only, 223 Arabidopsis cDNAs in 103 miRNA families and 197 members were finalized as potential miRNAs from 30,707 TAIR10 cDNAs (see System benchmarking and validation section for the detailed discussion).
Target identification pipeline
C-mii's target prediction menu consists of four submenus: sequence loading, target scanning, miRNA-target folding, and target annotation. Users may build a new project for target identification only or continue the project from the miRNA identification pipeline. Users may also upload a new set of nucleotide sequences in FASTA format or reuse the uploaded sequences from the miRNA identification process. However, the acceptable length of an input sequence for target identification is extended to 20,000 nt. Besides sequence uploading, users also need to select mature miRNAs of interest from miRBase and/or from predicted mature miRNAs saved from the miRNA identification pipeline. In our running example, 434 sequences previously reported as miRNA-specific targets in Arabidopsis were uploaded to a new project and all mature Arabidopsis miRNAs from miRBase were selected for target scanning.
Target scanning
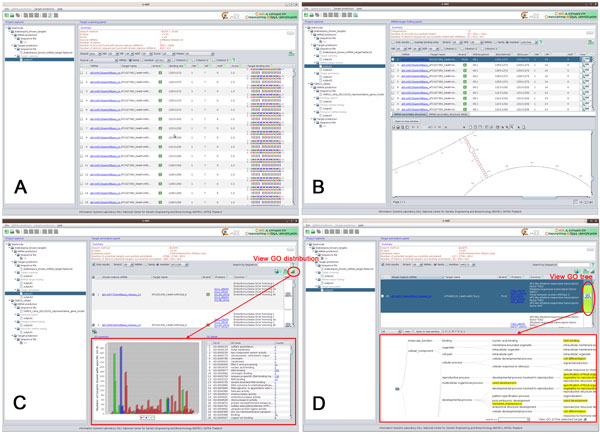
Based on plant-miRNA target binding through perfect or nearly-perfect complementarities [32,34], the target scanning module allows users to scan for complementary sites of selected mature miRNAs on input sequences. Based on criteria used in Rhoades et al. [33,35], users can customize the binding score as described in [38]. The scanning methods are the same as that of the homolog search with the reverse-complement of mature miRNAs used as queries for BLAST. Users can filter the results by the number of GU pairs, binding score, and mismatched position of interest. In addition, as the specific positions of mismatches affect miRNA targeting [22,36,37], C-mii also allows users to filter for target sequences whose miRNA binding site has no more than one mismatch at positions 1-9, no more than two consecutive mismatches, and no mismatches at positions 10 and 11. Figure 3A shows the target scanning results of our running example with default parameter settings. Using the plus strand only, 410 out of 434 sequences were identified as miRNA binding sites for 61 miRNA families and 133 members. All of them were selected as input for miRNA-target folding.
Figure 3.
Target identification results. (A) target scanning results, (B) miRNA-target folding results, (C) overall target annotation results with GO summary, and (D) individual target annotation result with GO tree.
MiRNA-target folding
The miRNA-target folding module helps users refine their scanning results. It uses UNAFold to predict the secondary structures of miRNA:target duplexes, MFEs for hybridization, and binding positions of miRNA-target pairs. Users can specify the temperature that might affect miRNA:target duplex formation. Figure 3B shows the miRNA-target folding results of our running example using the default 37 ºC. From the results, users can determine potential miRNA targets based on the binding score from target scanning, the number of mismatches and G:U pairs between miRNA and its potential target from miRNA-target folding, mismatched positions, MFEs, and overlapped binding positions between the two steps. In our example, with the plus strand only, all 410 sequences remained for target annotation.
Target annotation
The target annotation module supplies function and gene ontology (GO) for potential targets selected from the previous step. Users can choose a protein database and customize the E-value and number of hits for BLASTX. Figure 3C shows the target annotation results of 400 out of 410 sequences using default parameter settings (see System benchmarking and validation section for the detailed discussion). From the results, users may explore GO annotation for a set of targets. By clicking on "Graph icon," C-mii calculates and visualizes the distribution of selected targets' GO IDs, colored by GO molecular function (F), biological process (P), and cellular component (C). From this view, users may also explore potential targets annotated with the same GO. The "Go View" allows users to investigate GO annotation of an individual potential target via a GO tree (Figure 3D). Users may also follow web links to public databases of known miRNAs and target functions.
Summary views
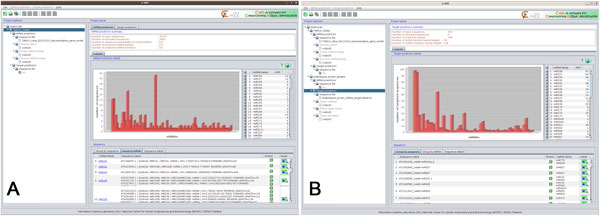
The project summary view provides users with the overall number of identified miRNA families and targets, which can be exported as a report and linked back to the detailed identification results. The miRNA prediction view (Figure 4A) shows the overall number of input sequences, excluded sequences, sequences potentially encoding miRNAs, and the identified miRNA families and members. The infographics highlight the distribution of the identified miRNA families. The "Group by sequence" and "Group by miRNA" options allow users to explore the identified miRNAs for the same sequence and the list of sequences identified for the same miRNA. The "Detail" icon allows users to follow the link back to the results of precursor miRNA folding. Similarly, the target prediction view (Figure 4B) shows the overall number of input sequences, excluded sequences, the identified targets, and source miRNA families and members having targets.
Figure 4.
Summary view. (A) miRNA prediction view, (B) target prediction view.
System validation and benchmarking
For system benchmarking, we applied C-mii to the four datasets: (i) TAIR10 cDNAs (33,602 sequences), (ii) TAIR10 miRNAs (176 sequences), (iii) Arabidopsis precursor miRNAs from miRBase 16 (213 sequences), and (iv) plant RNAs that are not miRNAs from Rfam 10 (16,219 sequences) (see Additional files 1, 2, 3, 4 for these sequences). The source of mature miRNAs was Arabidopsis miRNAs from miRBase 16. With default parameter settings except the E-value ≤ 1E-5 for BLASTX against the UniProt/Swiss-Prot protein database, Table 1 shows the number of remaining input sequences filtered for plus strand from each step of miRNA identification on the four datasets.
Table 1.
Number of remaining input sequences from each step of miRNA identification on the four datasets
| TAIR10 (cDNAs) | TAIR10 (miRNAs) | miRBase 16 (Arabidopsis only) | Rfam 10 (all plant RNAs except miRNAs) | |
|---|---|---|---|---|
| miRNA identification steps | 33,602 | 176 | 213 | 16,219 |
| 1. Sequence loading | 30,707 | 176 | 213 | 15,822 |
| 2. Homolog search | 1286 | 175 | 213 | 31 |
| 3. Primary miRNA folding | 567 | 173 | 209 | 0 |
| 4. Precursor miRNA folding | 223 | 164 | 195 | 0 |
Table 2 shows the number of true and false miRNAs identified by C-mii for the above datasets except for the TAIR10 miRNAs dataset, which is the subset of TAIR10 cDNAs. From the TAIR10 cDNA dataset, 164 out of 223 cDNAs selected by the precursor miRNA folding step were true positives (TP), consistently annotated as miRNAs in TAIR10. The remaining 59 cDNAs were considered false positives (FP). Twelve TAIR10 miRNAs were missing from the identification results and were considered as false negatives (FN) (see Additional file 5 for the list of these TP, FP, and FN of TAIR10 cDNAs). True negatives (TN) were the non-miRNA input sequences that were excluded from the miRNA identification results. With Arabidopsis precursor miRNAs from miRBase, 195 out of 213 miRNAs were correctly identified while the remaining 18 miRNAs were missing and considered as FN (see Additional file 6 for list of these FN). All plant RNAs that are not miRNAs from Rfam 10 were excluded from the identification results and considered as TN. The positive predictive value (PPV) for the TAIR10 cDNA dataset = 164/(164+59) = 0.7354 while the negative predictive value (NPV) = 30,472/(12 + 30,472) = 0.9996. The sensitivity of the TAIR10 cDNA dataset = 164/(164+12) = 0.9318 whereas the specificity = 30,472/(59 + 30,472) = 0.9981. The PPV, NPV, sensitivity, and specificity of the combined datasets were 0.8589, 0.9994, 0.9229, and 0.9987, respectively.
Table 2.
Number of TP, FP, FN, and TN of miRNA identification on the three datasets
| Data sets | Number of sequences | Number of identified miRNAs | TP | FP | FN | TN |
|---|---|---|---|---|---|---|
| TAIR10 cDNAs | 30,707 | 223 | 164 | 59 | 12 | 30,472 |
| miRBase 16.0 (Arabidopsis only) | 213 | 195 | 195 | 0 | 18 | 0 |
| Rfam 10 (all plant RNAs except miRNAs) | 15,822 | 0 | 0 | 0 | 0 | 15,822 |
| Total | 46,742 | 418 | 359 | 59 | 30 | 46,294 |
The previously reported 434 sequences of miRNA-associated targets of Arabidopsis (from 183 distinct TAIR10 gene loci and 49 Arabidopsis miRNA families) were used for benchmarking the target identification (see Additional file 7 for these sequences). The sensitivity was measured with two sets of parameter settings. The default settings included a binding score ≤ 4 in Target scanning, a folding temperature = 37 ºC for UNAFold in MiRNA-target folding, and an E-value ≤ 1E-20 for BLASTX against the plant-only UniProtKB/Swiss-Prot database in Target annotation. The customized settings used a binding score ≤ 6 in Target scanning and an E-value ≤ 1E-5 instead of 1E-20 in Target annotation. With mature Arabidopsis miRNAs from miRBase, C-mii identified 400 out of 434 miRNA-associated target sequences using default settings and the plus strand filter. Twenty-four sequences were lost due to the overly limited binding score, which was ≤ 4 in the default settings. The other ten miRNA-associated targets of seven distinct TAIR10 gene loci were lost in the annotation step; four out of seven were trans-acting siRNAs while the other three had too large hit E-values (> 0.1). Table 3 shows the number of known target sequences remaining from each step of target identification. With the use of the UniProtKB/Swiss-Prot protein database, the sensitivity of the identification calculated as TP/(TP + FN) was 0.922 and 0.933 for the default and customized settings respectively.
Table 3.
Number of remaining input sequences from each step of target identification on the previously reported miRNA-associated target sequences of Arabidopsis
| Step/Number of remaining sequences | Default settings | Customized settings | |
|---|---|---|---|
| Number of input sequences | 434 | 434 | |
| 1. Sequence loading | 434 | 434 | |
| 2. Target scanning* | 410 | 430 | |
| 3. miRNA-target folding | 410 | 430 | |
| 4. Target annotation** | UniProtKB/Swiss-Prot | 400 | 405 |
| UniProtKB/TrEMBL | 406 | 427 | |
* The default and customized binding scores for target scanning were ≤ 4 and ≤ 6, respectively.
** The default and customized BLASTX E-values for target annotation were 1E-20 and 1E-5, respectively. Both protein databases were prepared with plants only.
We measured the efficiency of multi-thread management on Ubuntu 9.10 (karmic) machine with four Intel(R) Core(TM)2 Quad CPU Q6600 at 2.4 GHz, 8GB RAM. The average speed of the miRNA and target identifications on TAIR10 cDNAs with default parameter settings was improved by 30 % and 46 % from single to two and four threads (see Table 4).
Table 4.
Time usage for each step of miRNA and target identifications on TAIR10 cDNA dataset with varied number of threads running
| 1 thread | 2 threads | 4 threads | |
|---|---|---|---|
| miRNA identification steps | |||
| Homolog search | 2:40:31 | 1:56:20 | 1:34:31 |
| Primary miRNA folding | 1:39:08 | 0:55:15 | 0:34:31 |
| Precursor miRNA folding | 0:18:24 | 0:22:10 | 0:17:40 |
| Target identification steps | |||
| Target scanning | 0:22:23 | 0:20:10 | 0:19:22 |
| miRA-target folding | 0:22:24 | 0:14:15 | 0:14:39 |
| Target annotation | 0:29:19 | 0:16:09 | 0:10:15 |
* The format of time usage is hours:minutes:seconds
Conclusions
This paper presents C-mii, a standalone software package for computational identification of plant miRNAs and targets. C-mii has been implemented as all-in-one Java package with following distinguishing features. First, it comes with graphical user interfaces of well-defined pipelines for both miRNA and target identifications, with reliable results. Second, it provides a set of filters allowing users to reduce the number of results corresponding to the recently proposed constraints in plant miRNA and target biogenesis. Third, it extends the standard computational steps of miRNA target identification with an miRNA-target folding module and GO annotation. Fourth, it supplies bird's eye views of the identification results with infographics and grouping information. Fifth, it provides helper functions for database update and auto-recovery to ease system usage and maintenance. Finally, it supports multi-project and multi-thread management to improve computational speed. With these features, C-mii is a very useful software package that can help accelerate the study of plant miRNAs and targets by plant biologists.
Availability and requirements
1) Project name: C-mii: A tool for plant miRNA and target identification
2) Project home page: http://www.biotec.or.th/isl/c-mii
3) Operating system(s): Windows and Ubuntu Linux 9.10 or higher
4) Programming language: Java and Python
5) Other requirements: -
6) License: GNU GPL
7) Any restrictions to use by non-academics: license needed
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
SN, WM, SI, and DW together designed software architecture and graphical user interfaces. SN developed Java-based interfaces and modules. WM implemented python scripts for results extractions. DW oversaw the software development. All authors helped test the overall functionalities of the system, and performed benchmarking and validation. SI helped draft the manuscript and DW wrote this manuscript. All co-authors read and approved the final manuscript.
Supplementary Material
FASTA (can be viewed with a text editor) - TAIR10 cDNA sequences. This file is used as input for benchmarking the miRNA identification pipeline. It is available under the Documentation & Benchmarking menu at http://www.biotec.or.th/isl/c-mii.
FASTA (can be viewed with a text editor) - TAIR10 miRNA sequences. This file is used as input for benchmarking the miRNA identification pipeline.
FASTA (can be viewed with a text editor) - Arabidopsis precursor miRNA sequences from miRBase 16. This file is used as input for benchmarking the miRNA identification pipeline.
(FASTA) - All plant RNA sequences that are not miRNAs from Rfam 10. This file is used as input for benchmarking the miRNA identification pipeline.
(Microsoft Excel) - List of true positives (TP), false positives (FP), and false negatives (FN) of the miRNA identification on TAIR10 cDNA dataset.
(Microsoft Excel) - List of false negatives (FN) of the miRNA identification on Arabidopsis precursor miRNAs from miRBase 16 dataset.
(FASTA) - Arabidopsis known miRNA target sequences. This file is used as input for benchmarking the target identification pipeline.
Contributor Information
Somrak Numnark, Email: somrak.num@biotec.or.th.
Wuttichai Mhuantong, Email: wuttichai.mhu@biotec.or.th.
Supawadee Ingsriswang, Email: supawadee@biotec.or.th.
Duangdao Wichadakul, Email: duangdao.wic@biotec.or.th.
Acknowledgements
The authors would like to thank Drs. Anan Jongkaewwattana, Samaporn Teeravechyan, and Sithichoke Tangphatsornruang for editing the manuscript and giving helpful comments on the results. This work was supported by grant FC0033 B21 (SPA B1-1) from Cluster Program Management Office (CPMO), National Science and Technology Development Agency (NSTDA), Thailand.
This article has been published as part of BMC Genomics Volume 13 Supplement 7, 2012: Eleventh International Conference on Bioinformatics (InCoB2012): Computational Biology. The full contents of the supplement are available online at http://www.biomedcentral.com/bmcgenomics/supplements/13/S7.
References
- Bartel DP. MicroRNAs. Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Zhang B, Pan X, Cobb GP, Anderson TA. Plant microRNA: a small regulatory molecule with big impact. Developmental Biology. 2006;289(1):3–16. doi: 10.1016/j.ydbio.2005.10.036. [DOI] [PubMed] [Google Scholar]
- Sunkar R, Chinnusamy V, Zhu J, Zhu J-K. Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends in Plant Science. 2007;12(7):301–309. doi: 10.1016/j.tplants.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Mallory AC, Vaucheret H. Functions of microRNAs and related small RNAs in plants. Nat Genet. 2006;38:S31–S36. doi: 10.1038/ng1791. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes ND, Freitas AT, Sagot M-F. Current tools for the identification of miRNA genes and their targets. Nucleic acids research. 2009;37(8):2419–2433. doi: 10.1093/nar/gkp145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dezulian T, Remmert M, Palatnik JF, Weigel D, Huson DH. Identification of plant microRNA homologs. Bioinformatics. 2006;22(3):359–360. doi: 10.1093/bioinformatics/bti802. [DOI] [PubMed] [Google Scholar]
- Zhang Y. miRU: an automated plant miRNA target prediction server. Nucleic acids research. 2005. pp. W701–704. [DOI] [PMC free article] [PubMed]
- Dai X, Zhao PX. psRNATarget: a plant small RNA target analysis server. Nucleic acids research. 2011. pp. W155–159. [DOI] [PMC free article] [PubMed]
- Bonnet E, He Y, Billiau K, Van de Peer Y. TAPIR, a web server for the prediction of plant microRNA targets, including target mimics. Bioinformatics. 2010;26(12):1566–1568. doi: 10.1093/bioinformatics/btq233. [DOI] [PubMed] [Google Scholar]
- Moxon S, Schwach F, Dalmay T, Maclean D, Studholme DJ, Moulton V. A toolkit for analysing large-scale plant small RNA datasets. Bioinformatics. 2008;24(19):2252–2253. doi: 10.1093/bioinformatics/btn428. [DOI] [PubMed] [Google Scholar]
- Milev I, Yahubyan G, Minkov I, Baev V. miRTour: plant miRNA and target prediction tool. Bioinformation. 2011;6(6):248–249. doi: 10.6026/97320630006248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F, Zhang B. Target-align: a tool for plant microRNA target identification. Bioinformatics (Oxford, England) 2010. [DOI] [PubMed]
- Wang X-J, Reyes J, Chua N-H, Gaasterland T. Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biology. 2004;5(9):R65. doi: 10.1186/gb-2004-5-9-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet E, Wuyts J, Rouze P, Van de Peer Y. Detection of 91 potential conserved plant microRNAs in Arabidopsis thaliana and Oryza sativa identifies important target genes. Proceedings of the National Academy of Sciences. 2004;101(31):11511–11516. doi: 10.1073/pnas.0404025101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Pan X, Wang QL, Cobb GP, Anderson TA. Identification and characterization of new plant microRNAs using EST analysis. Cell Res. 2005;15(5):336–360. doi: 10.1038/sj.cr.7290302. [DOI] [PubMed] [Google Scholar]
- Zhang B, Pan X, Cannon CH, Cobb GP, Anderson TA. Conservation and divergence of plant microRNA genes. The Plant Journal. 2006;46:243–259. doi: 10.1111/j.1365-313X.2006.02697.x. [DOI] [PubMed] [Google Scholar]
- Zhang B, Wang Q, Wang K, Pan X, Liu F, Guo T, Cobb GP, Anderson TA. Identification of cotton microRNAs and their targets. Gene. 2007;397:26–37. doi: 10.1016/j.gene.2007.03.020. [DOI] [PubMed] [Google Scholar]
- Xie FL, Huang SQ, Guo K, Xiang AL, Zhu YY, Nie L, Yang ZM. Computational identification of novel microRNAs and targets in Brassica napus. FEBS Letters. 2007;581(7):1464–1474. doi: 10.1016/j.febslet.2007.02.074. [DOI] [PubMed] [Google Scholar]
- Yin Z, Li C, Han X, Shen F. Identification of conserved microRNAs and their target genes in tomato (Lycopersicon esculentum) Gene. 2008;414:60–66. doi: 10.1016/j.gene.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Sunkar R, Jagadeeswaran G. In silico identification of conserved microRNAs in large number of diverse plant species. BMC Plant Biology. 2008;8(1):37. doi: 10.1186/1471-2229-8-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Pan X, Stellwag EJ. Identification of soybean microRNAs and their targets. Planta. 2008;229(1):161–182. doi: 10.1007/s00425-008-0818-x. [DOI] [PubMed] [Google Scholar]
- Frazier TP, Xie F, Freistaedter A, Burklew CE, Zhang B. Identification and characterization of microRNAs and their target genes in tobacco (Nicotiana tabacum) Planta. 2010;232(6):1289–1308. doi: 10.1007/s00425-010-1255-1. [DOI] [PubMed] [Google Scholar]
- Xie F, Frazier TP, Zhang B. Identification, characterization and expression analysis of MicroRNAs and their targets in the potato (Solanum tuberosum) Gene. 2011;473(1):8–22. doi: 10.1016/j.gene.2010.09.007. [DOI] [PubMed] [Google Scholar]
- Kim H-J, Baek K-H, Lee B-W, Choi D, Hur C-G. In silico identification and characterization of microRNAs and their putative target genes in Solanaceae plants. Genome. 2011;54(2):91–98. doi: 10.1139/G10-104. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Markham NR, Zuker M. In: Bioinformatics, Structure, Functions and Applications. Keith JM, editor. Vol. 2. Humana Press; 2008. UNAFold: software for nucleic acid folding and hybriziation; pp. 3–30. [Google Scholar]
- Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M. et al. A uniform system for microRNA annotation. Rna. 2003;9(3):277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang BH, Pan XP, Wang QL, Cobb GP, Anderson TA. Identification and characterization of new plant microRNAs using EST analysis. Cell Res. 2005;15(5):336–360. doi: 10.1038/sj.cr.7290302. [DOI] [PubMed] [Google Scholar]
- Zhang BH, Pan XP, Cox SB, Cobb GP, Anderson TA. Evidence that miRNAs are different from other RNAs. Cell Mol Life Sci. 2006;63(2):246–254. doi: 10.1007/s00018-005-5467-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Axtell MJ, Bartel B, Bartel DP, Baulcombe D, Bowman JL, Cao X, Carrington JC, Chen X, Green PJ. et al. Criteria for annotation of plant MicroRNAs. Plant Cell. 2008;20(12):3186–3190. doi: 10.1105/tpc.108.064311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297(5589):2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. Prediction of plant microRNA targets. Cell. 2002;110(4):513–520. doi: 10.1016/S0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes & Development. 2002;16(13):1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell. 2004;14(6):787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D. Specific effects of microRNAs on the plant transcriptome. Dev Cell. 2005;8(4):517–527. doi: 10.1016/j.devcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- Brodersen P, Voinnet O. Revisiting the principles of microRNA target recognition and mode of action. Nat Rev Mol Cell Biol. 2009;10(2):141–148. doi: 10.1038/nrm2619. [DOI] [PubMed] [Google Scholar]
- Fahlgren N, Carrington JC. miRNA target prediction in plants. Methods Mol Biol. 2010;592:51–57. doi: 10.1007/978-1-60327-005-2_4. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock R, van Dongen S. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic acids research. 2006;34(suppl 1):D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucl Acids Res. 2008;36(suppl_1):D154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A. Rfam: annotating non-coding RNAs in complete genomes. Nucleic acids research. 2005. pp. D121–124. [DOI] [PMC free article] [PubMed]
- The UniProt C. The universal protein resource (UniProt) Nucl Acids Res. 2008;36(suppl_1):D190–195. doi: 10.1093/nar/gkm895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghostscript. http://www.ghostscript.com/
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic acids research. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucl Acids Res %R 101093/nar/gkh340. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20(3):426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- The prefuse visualization toolkit. http://prefuse.org
- The ICEpdf Viewer. http://www.icesoft.org/projects/ICEpdf/overview.jsf
- The JFreeChart. http://www.jfree.org/jfreechart/
- Mhuantong W, Wichadakul D. MicroPC (uPC): a comprehensive resource for predicting and comparing plant microRNAs. BMC Genomics. 2009;10(1):366. doi: 10.1186/1471-2164-10-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FASTA (can be viewed with a text editor) - TAIR10 cDNA sequences. This file is used as input for benchmarking the miRNA identification pipeline. It is available under the Documentation & Benchmarking menu at http://www.biotec.or.th/isl/c-mii.
FASTA (can be viewed with a text editor) - TAIR10 miRNA sequences. This file is used as input for benchmarking the miRNA identification pipeline.
FASTA (can be viewed with a text editor) - Arabidopsis precursor miRNA sequences from miRBase 16. This file is used as input for benchmarking the miRNA identification pipeline.
(FASTA) - All plant RNA sequences that are not miRNAs from Rfam 10. This file is used as input for benchmarking the miRNA identification pipeline.
(Microsoft Excel) - List of true positives (TP), false positives (FP), and false negatives (FN) of the miRNA identification on TAIR10 cDNA dataset.
(Microsoft Excel) - List of false negatives (FN) of the miRNA identification on Arabidopsis precursor miRNAs from miRBase 16 dataset.
(FASTA) - Arabidopsis known miRNA target sequences. This file is used as input for benchmarking the target identification pipeline.