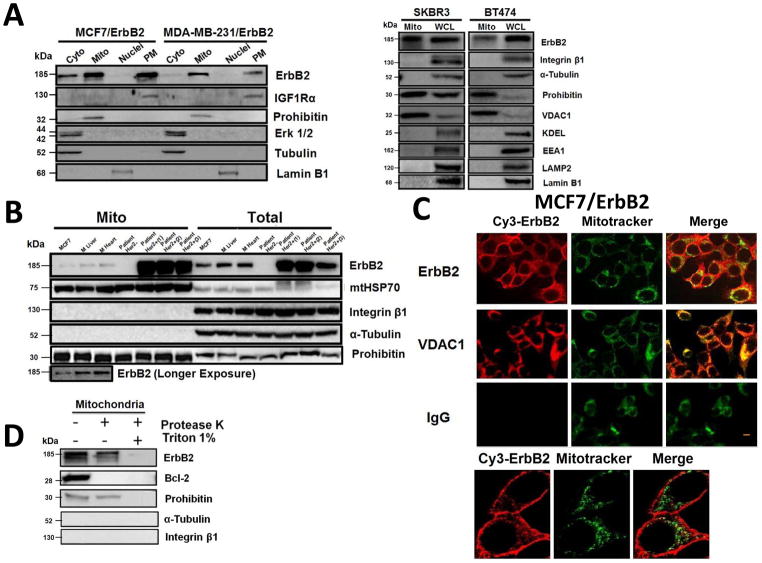
Fig. 1.
Localization of ErbB2 in mitochondria. (A) Cytosolic, nuclear, mitochondrial, and plasma membrane proteins were isolated and subjected to SDS-PAGE followed by probing with indicated antibodies. Two exogenous ErbB2 overexpressing breast cancer cell lines MCF7/ErbB2 and MDA-MB-231 (left), and two natural ErbB2-positive breast cancer cell lines SKBR3 and BT474 (right) were used for Western Blotting. VDAC1 and prohibitin were mitochondrial markers; Integrin β1 and IGF1Rα were plasma membrane markers; α-Tubulin and ERK were cytoplasmic markers; KDEL was an ER marker; EEA1 was an early endosomes marker; Golgi complex was a marker for the detection of Golgi; LAMP2 was an lysosome marker and Lamin B1 was a nucleus marker. (B) MCF7 cells, mouse heart and liver tissues, and ErbB2 positive and negative breast cancer patient samples were analyzed by Western blotting. (C) Co-localization of ErbB2 and mitochondria. Mitochondria were stained with Mitotracker-Green in ErbB2 transfected MCF7 cells. The cells were fixed and incubated with antibodies against ErbB2 (mouse), followed by incubation of monoclonal mouse Anti–Cy3 antibody (red). Images were analyzed with Nikon NIS-Elements AR software. Green: mitochondria; Red: ErbB2; Yellow: Co-localization of ErbB2 and mitochondria. The lower panel contains images with a higher magnification. Scale bars: 20 μm. (D) Localization of ErbB2 inside mitochondria. Intact mitochondria of SKBR3 cells were isolated and treated with Protease K at 0.5 mg/ml for 15 min at room temperature. 1% Triton was added into the reaction and 2 mM PMSF was added to stop the reaction. Each reaction was started with an equal amount/volume of mitochondria lysate. Mitochondrial lysate then was loaded onto 8% SDS PAGE followed by blotting with anti-ErbB2, anti-BCL-2 and anti-Prohibitin antibodies.