Fig. 5.
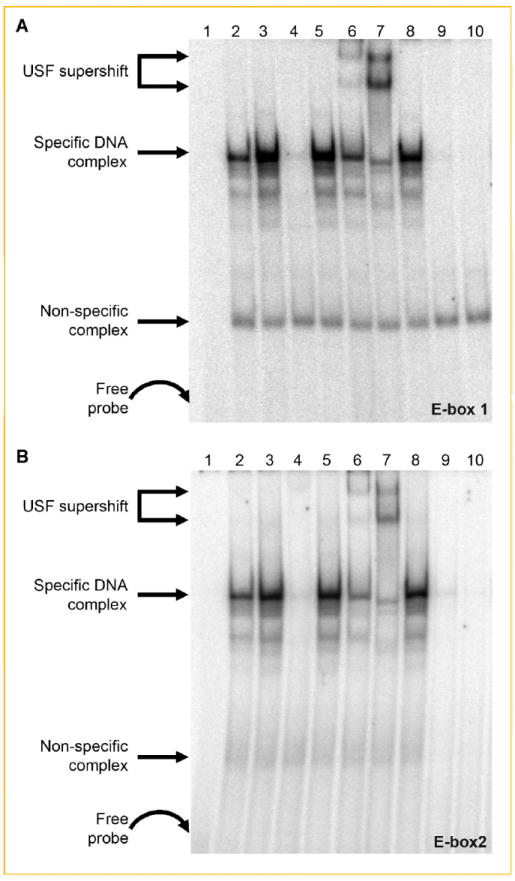
Identification of USF-1 and USF-2 as E-box-binding proteins. Nuclear proteins from untreated and Quercetin-treated cells were incubated with 32P-labeled double-stranded oligonucleotides to E-box1 (A) or E-box2 (B). Lane 1, probe only; lane 2, untreated NE; lane 3, quercetin-treated NE; lane 4, quercetin-treated NE and 100-fold excess of unlabeled probe; lane 5, quercetin-treated NE and 100-fold excess of mutated E-box1 (A) or E-box2 (B); lane 6, quercetin-treated NE and USF-1 antibody; lane 7, quercetin-treated NE and USF-2 antibody; lane 8, quercetin-treated NE and c-Myc anibody; lane 9, unstimulated NE treated with lambda protein phosphatase; and lane 10, quercetin-stimulated NE treated with phosphatase. The two supershifted bands in (A) and (B) lane 7 and, to a lesser extent, lane 6 likely are due to the relative abundance of different complexes (i.e., USF-2 homodimers vs. USF-1/2 heterodimers) that bind to E-box1 and E-box2 as a function of stimulus conditions [e.g., Allen et al., 2005; Qi et al., 2006].
