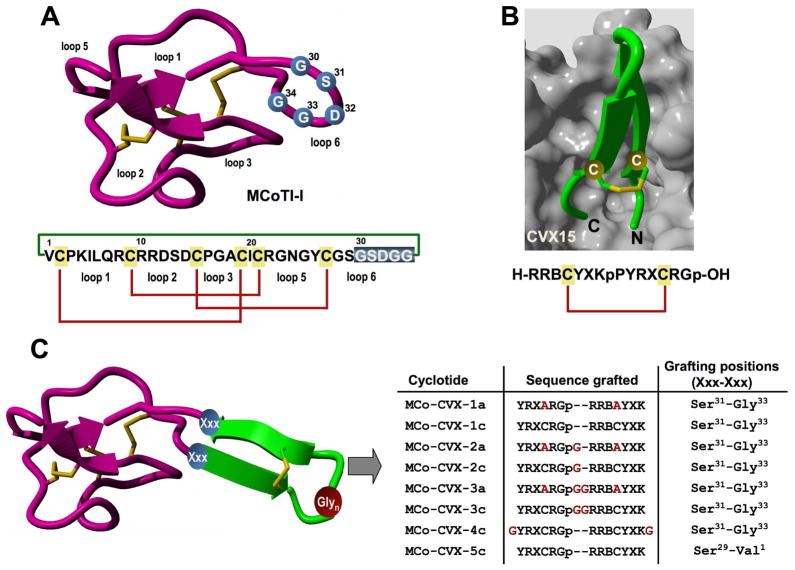
Figure 1.
Design of MCoTI-based cyclotides to target the cytokine receptor CXCR4. A. Primary and tertiary structures of cyclotide MCoTI-I. Structure is based on a homology model using the solution structure of MCoTI-II as template (PDB: 1IB9).45 The backbone cyclized peptide (connecting bond shown in green) is stabilized by the three-disulfide bonds (shown in red). The residues used for the grafting of a CVX15-based peptide are shown in blue on the structure and sequence of MCoTI-I. B. Sequence and co-crystal structure of peptide CVX15 bound to cytokine receptor CXCR4 (PDB: 3OE0).40 Peptide CVX15 is shown as a ribbon representation in green with the side-chains of the Cys residues involved in the disulfide bond in ball-and-stick form. The solvent accessible surface of the binding site of CXCR4 is shown in grey. C. Scheme depicting the approach used to design the different MCo-CVX cyclotides. A circularly permuted version of CVX15 was grafted onto loop 6 of MCoTI-I at different residues. The CVX15-based insert was created by joining the C and N-terminus directly through a flexible Glyn linker and opening the new sequence at the D-Pro-Pro segment. Residues in red denote mutations or extra Gly residues introduced to increase flexibility. Single letter codes B, X and p represent the amino acid, 2-naphthylalanine, citruline and D-proline, respectively. Molecular graphics were built with Yasara (www.yasara.org).