Figure 1.
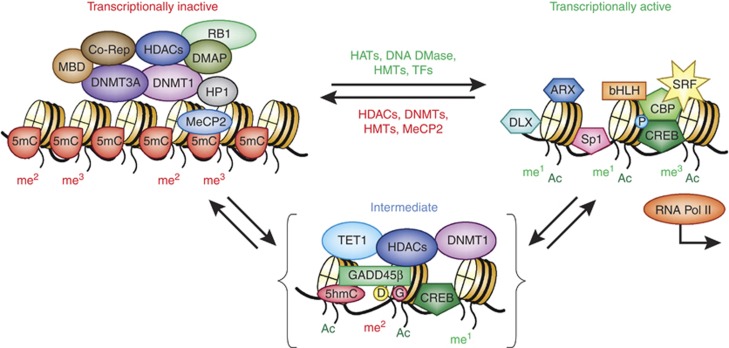
Proteins bound to DNA and histones cooperate in facilitating transitions between active and inactive chromatin states. Schematic representation of the transitions between a transcriptionally inactive promoter (left) and a transcriptionally active state (right). The transcriptionally inactive state is characterized by DNA methylation and the binding of various repressor proteins, including DNA methyltransferase 1 (DNMT1) and 3A, methyl-binding domain proteins (MBDs, MeCP2), co-repressors, and modified histones associated with repressive chromatin marks (H3K9me2, H3K9me3, H3K27me2, H3K27me3, etc.). The intermediate state (shown in brackets) is stable and ‘poised' for either repression or activation. In the intermediate state, the DNA/protein complex is characterized by the binding of DNMT1 to unmethylated CpGs and ten-eleven translocase-1 (TET-1) bound to 5-methylcytosines (5mCs) and 5-hyroxymethylcytosine (5hmCs). In the transitional phase, DNMT1 is associated with histone deacetylases (HDACs) and excess DNMT3A shifts this towards the inactive state (left). The binding of TET1 to hydroxymethylated CpGs in this same intermediate state reinforces stable repression until the entry of GADD45β, which recruits proteins required for DNA demethylation (deaminases and glycosylases). DNA demethylation is accompanied by additional histone modifications (mediated by HATs and HMTs). Hydroxymethylated CpGs are further modified and removed. In this model, HDAC inhibitors facilitate a disruption of the inactive state and depending on the availability of GADD45β, DNA demethylation ensues (Kundakovic et al, 2009; Guidotti et al, 2011). In the active (open) state, various transcription factors (TFs) bind and occupy their specific DNA recognition sites enabling transcription. The specific TFs involved depend on the gene being activated and the neuronal phenotype (West and Greenberg, 2011). Some of the transcription factors are shown bound to the intermediate state (such as CREB, which upon phosphorylation (P) recruits the histone acetyltransferase CBP). Transcriptionally active promoters are represented as an open chromatin structure characterized by the presence of acetylated (H3K9ac, H3K14ac) and methylated (eg, H3K4me1, H3K4me3, H3K9me1, H3K27me1, H3K79me1, etc) histones. The model highlights repressive roles for DNMT1 and TET1, which depends upon the availability of accessory proteins (DNMT3A and GADD45β, respectively) to modify their function in postmitotic neurons. Based on localization studies of DNMT1 in GABAergic neurons (Kadriu et al, 2011) and GADD45β in pyramidal neurons (Gavin et al, 2012), these mechanisms are likely unique to specific types of neurons depending on neurotransmitter phenotype. ARX, aristaless-related homeobox; bHLH, basic helix-loop-helix transcription factors; CBP, CREB-binding protein; Co-Rep, co-repressor proteins; CREB, cyclic AMP response element-binding protein; D, deaminase; DLX, distal-less homeobox; DMAP1, DNA methyltransferase 1-associated protein; DNA DMase, DNA demethylase; G, gycosylase; HATs, Histone acetyl transferases; HMTs, histone methyl transferases; HP1, hetrochromatin protein 1; me1, monomethyl; me2, dimethyl; me3, trimethyl; MeCP2, methyl CpG-binding protein 2; P, phosphoryl group; RB1, retinoblastoma 1; SP1, promoter-specific transcription factor; SRF, serum response factor; TFs, transcription factors.