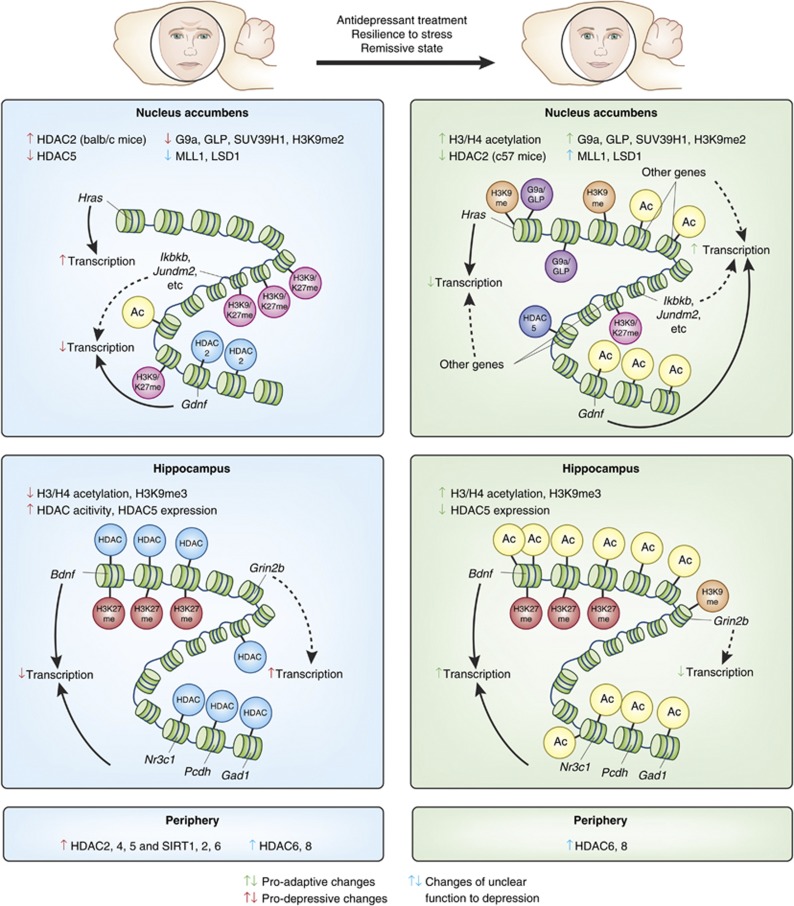
Figure 3.
Epigenetic changes in the depressed brain. Summary of epigenetic changes reported to date in the brain and periphery in depressed humans or after chronic stress in animal models, as well as after antidepressant treatment (or in resilient or remitted subjects). Global as well as representative promoter-specific changes are shown. Changes marked by green arrows are thought to be adaptive/antidepressant/pro-resilient. Changes marked by red arrows are thought to be pro-depressive. Changes marked by blue arrows have as yet an unclear role in depression. Solid-line arrows represent established connections, whereas dotted-line arrows represent putative connections. In the nucleus accumbens (NAc) of a stress-susceptible mouse strain (ie, balb/c), global HDAC2 expression and HDAC2 binding on specific genes (eg, Gdnf) were increased after chronic stress (Uchida et al, 2011). These changes reversed by antidepressant treatment, while the opposite adaptive changes were observed in a more resilient stain of mice (c57b1/6) and in humans (Covington et al, 2009). On the contrary, HDAC5, a class II HDAC, were decreased in animals susceptible to stress and was increased after antidepressant treatment (Renthal et al, 2007). These results suggest that there may be two distinct groups of pro-resilient/antidepressant genes: those targeted by HDAC5 and that show decreased acetylation (and possibly decreased transcription), and those targeted by HDAC2 and that show increased acetylation (and possibly increased transcription). In addition, global levels of repressive histone methylation machinery for H3K9 (G9a, GLP, and SUV39H1), which presumably control global H3K9me2 levels, as well as H3K9me2 binding on specific genes (eg. Hras), were decreased in the NAc of susceptible animals after chronic social defeat stress, while the opposite changes were observed in resilient animals (Covington et al, 2011a). Also, histone methyltransferase (MLL1) and demethylase (LSD1) for H3K4 were decreased in susceptible animals, while they were increased in resilient animals after social defeat, with no change in global levels of H3K4 methylation (Covington et al, 2011a). The role of these changes are yet unclear. A ChIP-chip study has revealed dynamic changes in repressive histone methylation (H3K9/K27 methlation) in the upstream regulatory regions genome-wide after social defeat, with more genes showing increased methylation (eg, Ikbkb and Jundm2) and putative decreased transcription (Wilkinson et al, 2009). Many of these changes were reversed by antidepressant treatment or were not observed in resilient animals. In the hippocampus of susceptible rodents after various stress paradigms, global acetylation levels on numerous histone residues were decreased, accompanied by increased HDAC expression or activity (Covington et al, 2011b; Hollis et al, 2011; Ferland and Schrader, 2011). These changes were either not observed, returned to baseline, or changed in the opposite direction in resilient animals or after antidepressant treatment. Similar changes were observed for H3K9me3 in the hippocampus (Hunter et al, 2009). Gene-specific epigenetic changes were also observed in the hippocampus (eg, Bdnf, Nr3c1 gene cluster, and Grin2b). In the peripheral blood of depressed patients, HDAC2,4, and 5, as well as SIRT1, 2, and 6 were increased only in the depressive and not in the remissive state, while HDAC6 and 8 were increased in all patients regardless of states (Iga et al, 2007; Hobara et al, 2010; Abe et al, 2011).