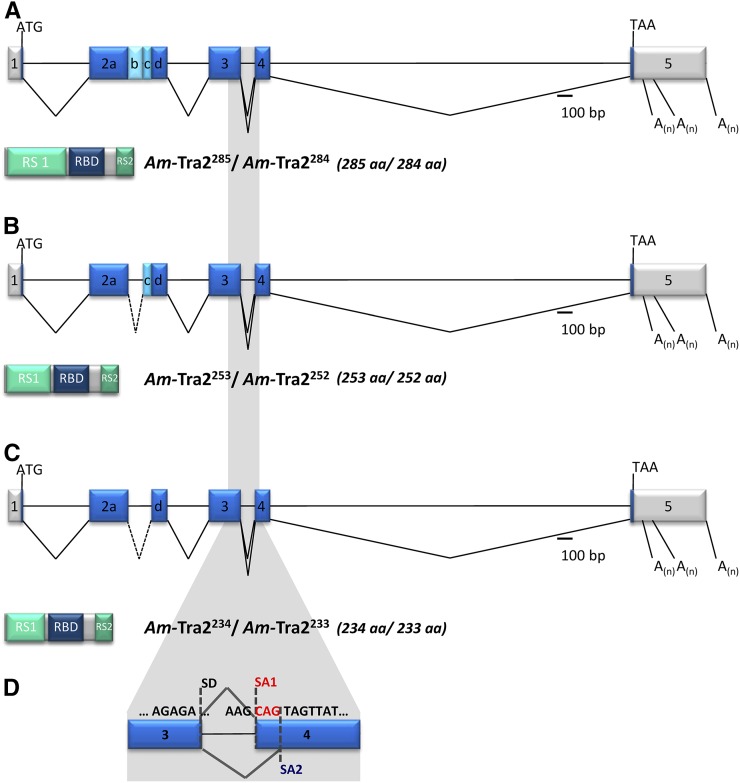
Figure 1 .
Genomic organization of the Am-tra2 gene and the alternatively spliced mRNAs. Schematic representation of the intron and exon organization (presented as lines and boxes, respectively). The alternatively spliced transcripts are indicated by the connecting lines between exons. The three alternative polyadenylation sites are labeled “A(n).” The scale denotes the relative size of the introns and exons. The 5′- and 3′-UTRs are presented in gray and the ORF in blue boxes. Below the genomic organization, the domain structure and relative size of the predicted Am-Tra2 proteins are shown (RS, arginine/serine-rich domain; RBD, RNA-binding domain). Superscript of Am-Tra2 proteins denotes the number of amino acids in that particular protein isoform. (A) Am-Tra2285 and Am-Tra2284. (B) Am-Tra2253 and Am-Tra2252. (C) Am-Tra2234 and Am-Tra2233. (D) Alternatively spliced variants in exon 4 producing trinucleotide and single-amino-acid differences. This splicing affected all three transcripts shown in A–C that are denoted as the Am-tra2284, Am-tra2252, and Am-tra2233 transcripts. SA1 and SA2 label the alternatively splice acceptors in exon 4.