Figure 2.
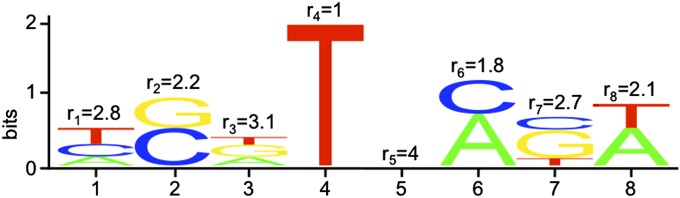
The rate of deleterious mutations at a binding site can be calculated directly from a PWM. An arbitrary PWM logo is shown, for a binding site of length n = 8. The x-axis shows nucleotide position, and the y-axis shows the total information contained in that nucleotide. The relative heights of the different letters (A, C, G, or T, corresponding to the different DNA bases) correspond to the relative frequency with which they occur in functional binding sites. The value of ri at each position gives the degeneracy, i.e., the average number of different bases that occur at that nucleotide in a functional binding site, calculated from the relative frequency with which each base is found in a functional site at that position. A deleterious mutation occurs when a disallowed nucleotide occurs at one of these positions. The total rate of deleterious mutations for the PWM, u−, can be determined by counting the total number of deleterious mutations that can occur at each nucleotide; i.e., , where is the average degeneracy across the PWM.