Table 3. Posterior means and 95% highest posterior density intervals (HPD; in parenthesis) for heritability estimates, genetic and phenotypic correlations obtained with the four models considered in the study and mean correlation between individual EBVs obtained from five sub-dataset samples for each model.
| Item | Model | |||
|---|---|---|---|---|
| |
M1 |
M2 |
M3 |
M4 |
| Heritabilitya | ||||
| h2D | 0.122 (0.094 to 0.150) | 0.099 (0.073 to 0.125) | 0.381 (0.330 to 0.433) | 0.265 (0.214 to 0.317) |
| h2C | — | — | 0.335 (0.297 to 0.737) | 0.215 (0.172 to 0.259) |
| h2TOT | 0.122 (0.094 to 0.150) | 0.099 (0.073 to 0.125) | 0.005 (−0.005 to 0.015) | 0.014 (−0.006 to 0.033) |
| Correlationb | ||||
| raD,C | — | — | −0.995 (−1.007 to −0.983) | −0.976(−1.018 to −0.938) |
| rPeD,C | — | −0.259 (−0.279 to −0.239) | −0.276(−0.296 to −0.256) | −0.279 (−0.298 to −0.260) |
| rEBVc | 0.746 | 0.726 | 0.847 | 0.855 |
h2D is the direct heritability for social dominance (measured as a ratio of direct variance on the total phenotype, as 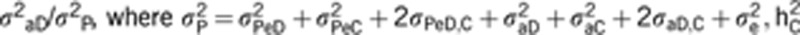 is the indirect heritability
is the indirect heritability 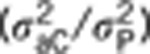 , and h2TOT is the TBV computed as
, and h2TOT is the TBV computed as 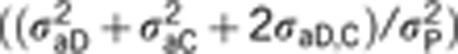
raD,C is the genetic correlation for direct and indirect effects (that is, σaD,C/(σ2aDσ2aC)0.5), rPeD,C is the phenotypic correlation for the same components, calculated as 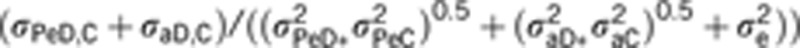
rEBV is the average correlation between direct EBVs estimated from five sub-datasets (that is, 13 610±236 data) obtained after retaining about half of the dataset (that is, 5452±33 cows). ±Correlations were carried out on the EBV of individuals shared by each sub-dataset (that is, 2902±36 cows) and were averaged for each model.
