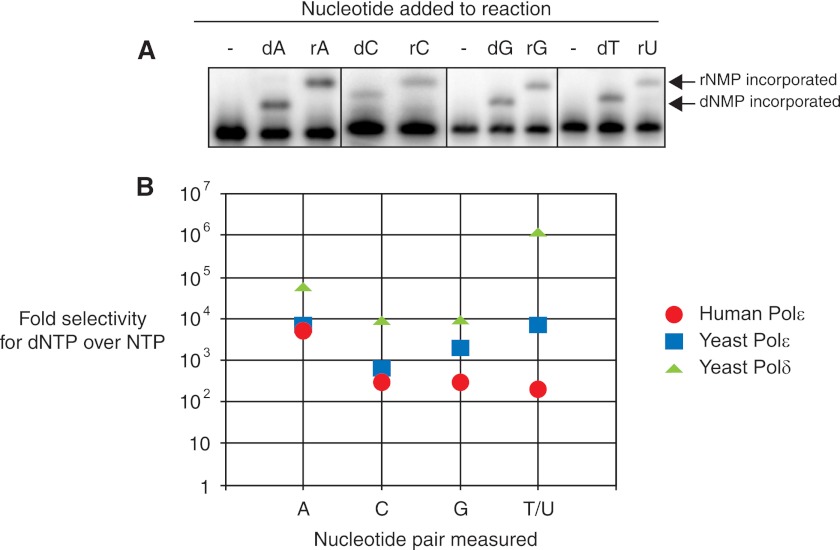
FIGURE 2.
Ribonucleotide selectivity of human Pol ϵ. A, human Pol ϵ was incubated with 100 nm primer-template substrate where the next correctly base-paired incoming nucleotide is indicated. The indicated nucleotide was added at its physiological concentration (Table 1). The amount of Pol ϵ was adjusted until less than 20% of the primer was extended. For each dNTP, 0.3 nm Pol ϵ was used. For ribonucleotide incorporation, the following concentrations of Pol ϵ were used: rATP, 12 nm; rCTP, 6 nm; rGTP, 3 nm; rUTP, 0.6 nm. Shown are individual pairs of deoxyribo- and ribonucleotide for comparison. Arrows indicate the migration of the single incorporated dNMP or rNMP product. B, ribonucleotide selectivity values from experiments in A were calculated using Equation 1 (described under “Experimental Procedures” and in Ref. 5) and then plotted for human Pol ϵ as the -fold selectivity of dNTP over NTP (red circles). Ribonucleotide selectivity values for yeast Pol ϵ (blue squares) and yeast Pol δ (green triangles) are shown for comparison and were taken from Ref. 5.