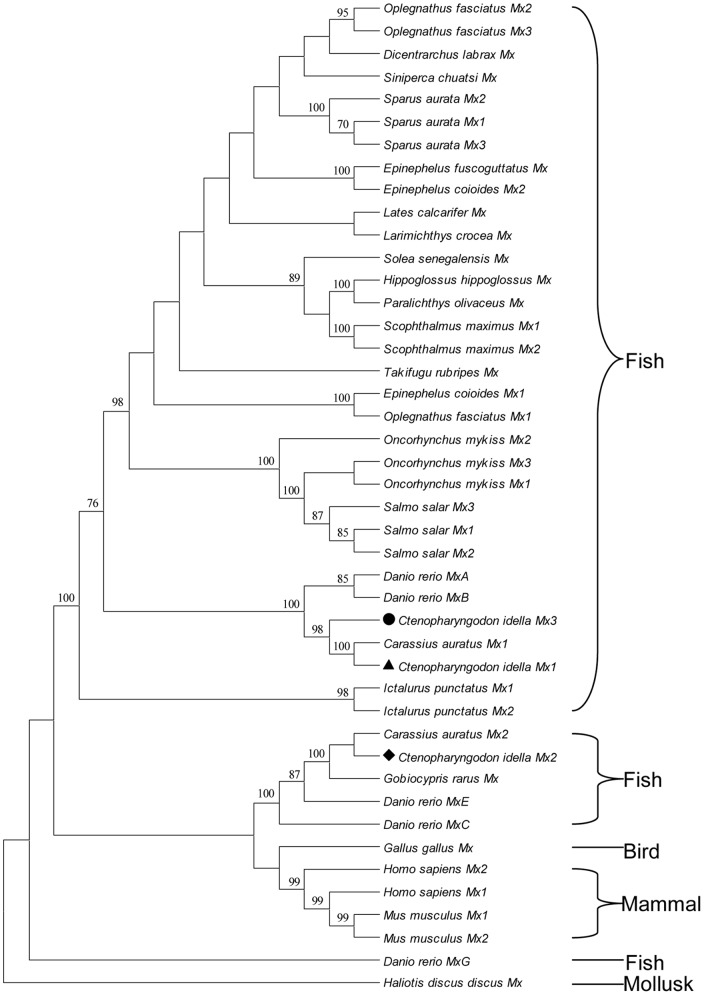
Figure 1. Phylogenetic relationships of all the fish, chicken, mouse and human Mx protein sequences in GenBank.
(Danio rerio MxD and MxF sequences are not included due to just partial sequences available). Maximum-likelihood phylogenetic tree generated from a MAFT alignment and MEGA 5.1 program. Haliotis discus discus Mx was employed as the outgroup. The bar indicates the distance. CiMx1, CiMx2 and CiMx3 were marked with triangle (▴), diamond (♦) and circle (•), respectively. The protein IDs are as follows: Ctenopharyngodon idella Mx1 ADU33870, Mx2 AAQ95584, Mx3 ADZ44601; Carassius auratus Mx1 AAP68828, Mx2 AAP68827; Danio rerio MxA NP_891987, MxB Q800G8, MxC NP_001007285, MxE NP_878287, MxG CAD67761; Dicentrarchus labrax Mx AAR99718; Epinephelus fuscoguttatus Mx ADE80885; Epinephelus coioides Mx1 ABD95979, Mx2 ABD95982; Gobiocypris rarus Mx ABL61237; Gallus gallus Mx CAA80686; Homo sapiens Mx1 NP_001171517, Mx2 NP_002454; Hippoglossus hippoglossus Mx AAF66055; Ictalurus punctatus Mx1 Q7T2P0, Mx2 AAY33864; Lates calcarifer Mx AAW22002; Larimichthys crocea Mx ABJ56003; Mus musculus Mx1 NP_034976, Mx2 NP_038634; Oplegnathus fasciatus Mx1 ACF75866, Mx2 ACF75867, Mx3 ACF75868; Oncorhynchus mykiss Mx1 AAA87839, Mx2 AAC60214, Mx3 AAC60215; Paralichthys olivaceus Mx BAC76769; Scophthalmus maximus Mx1 AAT57877, Mx2 AAT57878; Sparus aurata Mx1 ACK99554, Mx2 ACK99553, Mx3 ACN22085; Salmo salar Mx1 AAB40994, Mx2 AAB40995, Mx3 AAB40996; Solea senegalensis Mx AAV49303; Siniperca chuatsi Mx AAQ91382; Takifugu rubripes Mx AAO37934; Haliotis discus discus Mx ABI53802.