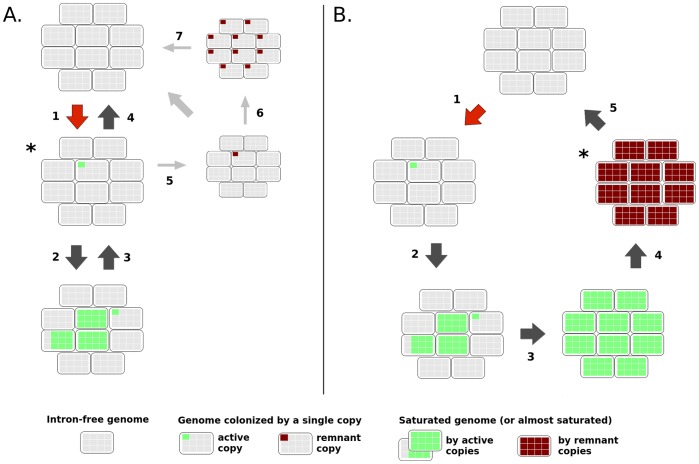
Figure 1. Two proposed extinction-recolonization dynamics of group II introns in a bacterial population.
(A) Selection-driven extinction (Sel-DE) model: acquisition of a functional copy via a MGE-mediated horizontal transfer (step 1, red arrow), quick element proliferation in the host genome and in the population (step 2), and removal of bacterial individuals with proliferated copies through purifying selection (step 3). Individuals with few copies ultimately disappear from the population through rampant purifying selection or eventually genetic drift, resulting in complete loss of the group II intron from the bacterial population (step 4). Alternatively, not proliferated copies may be inactivated by mutation (step 5), evolve as neutral genomic regions and eventually become fixed in the population (step 6). Residual inactivated copies then undergo slow degradation until complete loss from the bacterial genome (step 7). (B) Saturation-driven extinction (Sat-DE) model: the first two steps are identical to those of the Sel-DE model, but proliferation results in the saturation of all available homing sites in the population (step 3). Proliferated copies then start to degenerate (step 4) and are slowly removed from the population through random mutations and deletions (step 5). Bacterial populations are represented by sets of cells (rounded rectangles) harboring available homing sites (grey boxes) or homing sites occupied by active (green boxes) or inactive intron copies (dark red boxes). The starred state in each model is those predicted to be the most frequently observed when a group II intron copy is present. Smaller cartoon sizes (in A) reflect infrequent alternative.