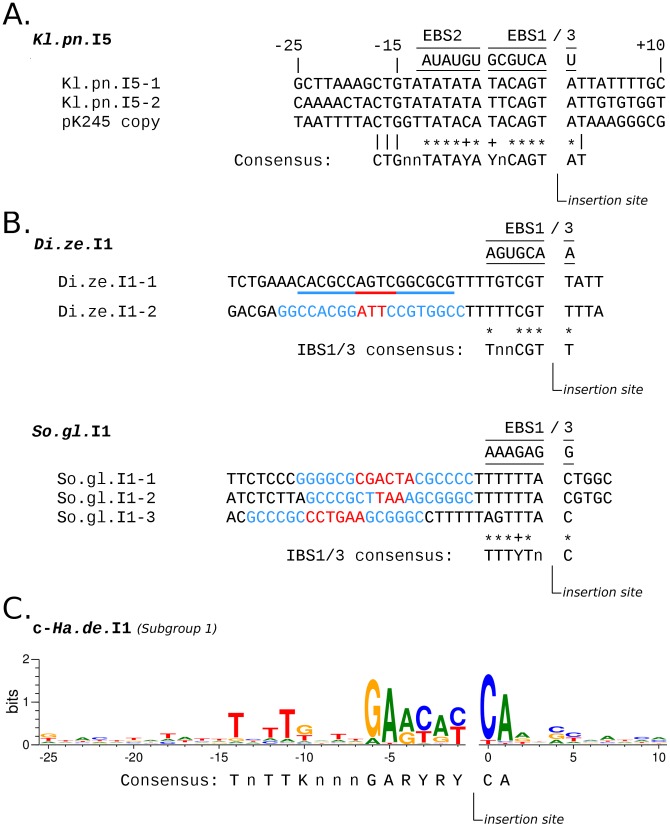
Figure 3. Inferred homing sites for Kl.pn.I5, Di.ze.I1, So.gl.I1, and c-Ha.de.I1 introns.
(A) Sequence spanning from positions −25 to +10 relative to full-length Kl.pn.I5 copy insertion sites are displayed and compared to predicted EBS. Identical bases and those which fit the Watson-Crick (stars) or the Wobble (plus) base-pairing with EBS motifs were used to construct the consensus homing site. (B) Regions surrounding Di.ze.I1 and So.gl.I1 full-length copies are displayed and compared to predicted EBS1 and EBS3. Most represented nucleotides at a given position and which fit the Watson-Crick (stars) or the Wobble (plus) base-pairing with EBS motifs were used to construct the consensus homing site. Stem-loops belonging to Rho-independent transcription terminators inferred by ARNold [58] are colored in blue (stems) and red (loops). Underlined sequences are stem-loops not detected by ARNold. (C) Weblogo diagram of the −25 to +10 region surrounding the 26 full-length c-Ha.de.I1 copies belonging to subgroup 1. Bases which show relative conservation were used to construct the consensus homing site.