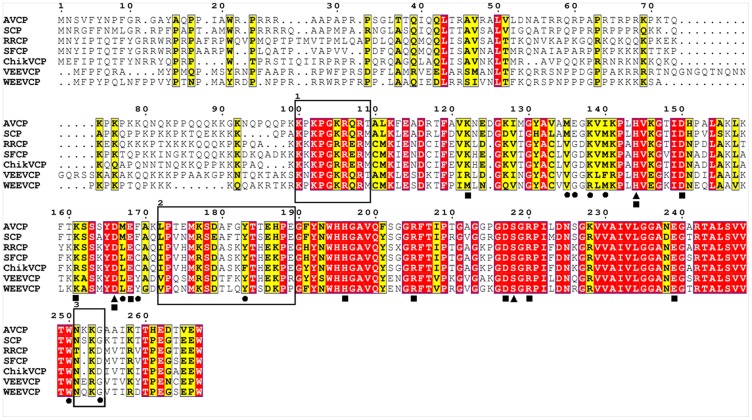
Figure 2. Multiple sequence alignment of AVCP with CPs from other alphaviruses.
The conserved residues are shown in red background. The circles under the amino acids indicate the hydrophobic pocket residues interacting with the dioxane molecule while the triangles denote the catalytic triad residues. The residues forming salt bridges are denoted by squares below the residues. The motif responsible for specificity in encapsidation is highlighted in rectangular box 1. Interdomain flexible loop residues are shown in box 2 whereas the flexible loop separating the two pockets for E1 and E2 binding is shown in box 3. Capsid protein sequences used for alignment are: Aura virus (AVCP), Sindbis (SCP), Ross River (RRCP), Semliki Forest (SFCP), Chikungunya (ChikVCP), Venezuelan equine encephalitis (VEEVCP) and Western equine encephalitis (WEEVCP).