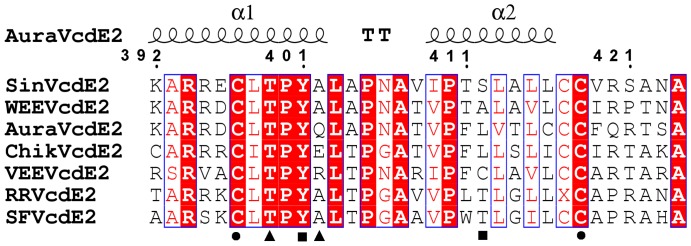
Figure 5. Multiple sequence alignment of the cytoplasmic domain of E2 (cdE2) from different alphaviruses.
The secondary structure of 33 residues from Aura virus cdE2 displays a helix-loop-helix structure. Highly conserved residues are in red background. The circles under residues denote the disulfide bridge forming Cys residues, squares indicate the residues responsible for polar interactions within the E2 helix-loop-helix motif and the triangles represent the residues of E2 making polar interactions with CP. cdE2 sequences used for alignment are: Sindbis (SinVcdE2), Western equine encephalitis (WEEVcdE2), Aura (AuraVcdE2), Chikungunya (ChikVcdE2), Venezuelan equine encephalitis (VEEVcdE2), Ross River (RRVcdE2) and Semliki Forest (SFVcdE2).