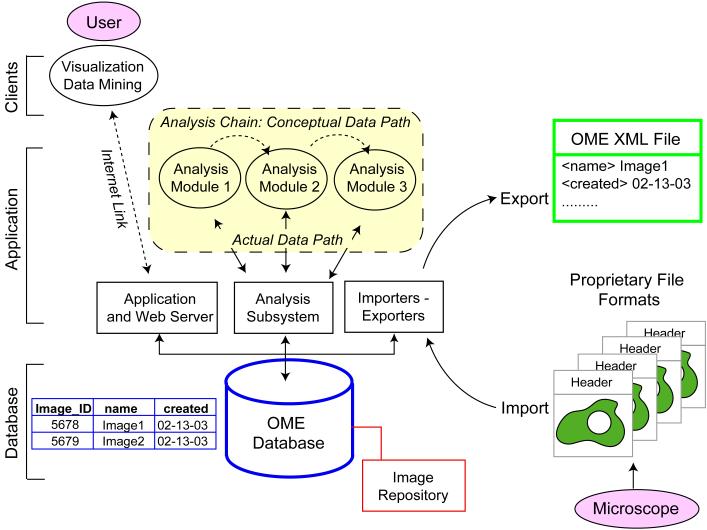
Figure 2. The Database is the Interface: OME Architecture.
OME is constructed as a standard “three-tier” application with a relational database that stores information in a table-based structure (blue), an application server that processes data and a client that lives on the users desktop and communicates via the internet (that is, via IP). Multiple clients can communicate with OME including Web browsers, commercial microscopy software and data mining applications. The OME data model is instantiated via a relational database (“OME database;” blue) in which metadata is stored in tables as specified by the schema and binary image data is stored in a trusted file system (the “image repository;” red). When data is transported between databases, or stored in a flat file, metadata and image data in the database are translated into XML (“OME XML File;” green). The OME database communicates with analysis modules via a subsystem (“analysis subsystem”) that ensures the consistent treatment of semantic datatypes. The analysis modules also calculate and store the history of the analysis chain (see online supplemental material). When analysis modules are chained together, each communicates independently with the database (“actual data path;” yellow block) even though the conceptual path appears is from one module to the next.(“conceptual data path”). Existing commercial or independent software tools can read OME data without substantial modification. OME itself is open source and available through a LGPL license but applications that talk to it can be either open or proprietary.