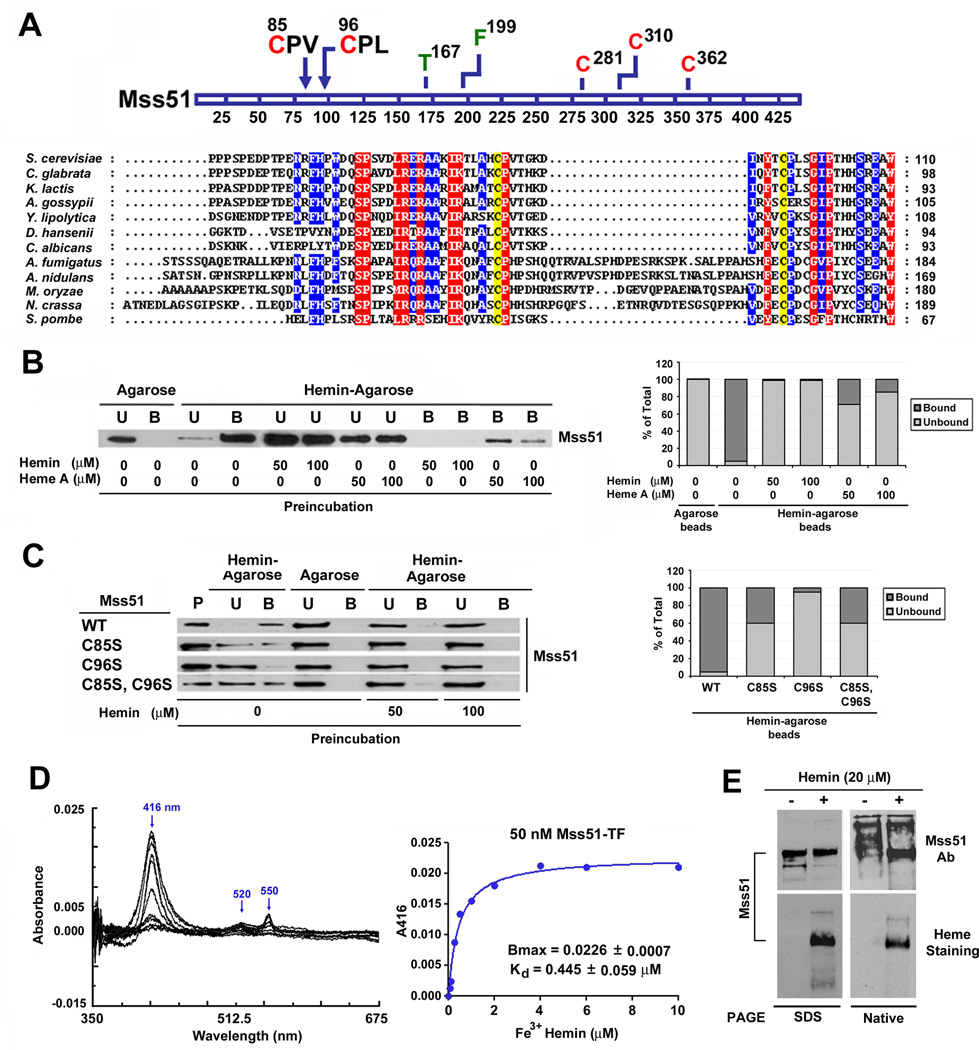
Figure 1. Mss51 contains two conserved CPX motifs and binds heme in vitro.
(A) Partial N-terminal sequence alignment of Mss51 from 12 fungal species. Sequences were aligned using the CLUSTALW algorithm (Blosum62 scoring matrix) in Bioedit software. Amino-acids that are conserved in all sequences are shaded red and those conserved in at least 9 of 12 sequences are shaded blue. The conservation of two cysteines in the CPX motifs is marked in yellow. In the linear graph of the protein, all the cysteines (red) and other relevant residues for this study (green or black) are indicated.
(B and C) Immunoblot analyses of the binding to hemin-agarose beads of recombinant (B) wild-type Mss51 or (C) CPX mutant Mss51. Mss51 was pre-incubated or not with the indicated concentrations of either hemin or heme A prior to incubation with the beads. To quantify the signals, the images were digitalized and densitometric analyses performed using the histogram function of the Adobe Photoshop program. The graph represents the proportion of bound versus unbound protein for each sample. B, bound; U, unbound; P, protein alone.
(D) Difference spectroscopy titration of heme binding to Mss51. Difference absorption spectra and titration curves of wild-type Mss51 (50 nM) with increasing concentrations of Fe3+ hemin (from 0 to 10 µM) as indicated. The curves were generated from fits to an equation describing a single binding site using GraphPad Prism.
(E) Analyses of Mss51 and heme-bound-Mss51 by SDS- and Native-PAGE. The upper panel represents immunoblot analyses with an anti-Mss51 antibody. The lower panel represents in gel heme staining.