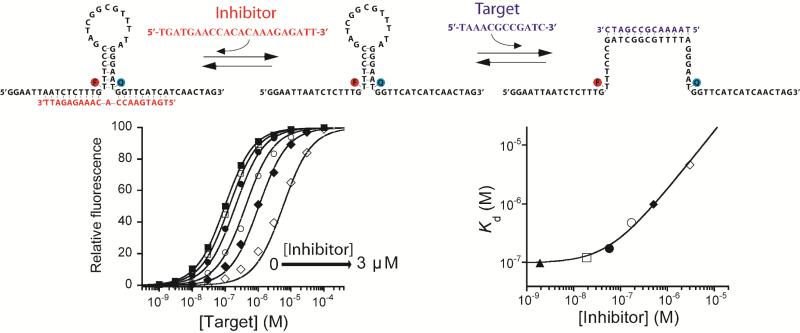
Figure 2. Rational design of an allosterically inhibited biosensor.
(Top) We have engineered allosteric inhibition into a molecular beacon by adding two single-stranded “tails” that, together, serve as the allosteric binding site. The binding of an inhibitor that bridges across these tails stabilizes the non-binding state of the beacon, consequently reducing target affinity and pushing the useful dynamic range to higher target concentrations. (Bottom) As expected, the extent to which this allosteric inhibition shifts the dynamic range of the beacon (here and in all the experiments at 10 nM concentration) can be fine-tuned by varying the inhibitor concentration. Here we have employed inhibitor concentrations producing (from left to right) 0, 5, 25, 50, 75, 90 and 98% occupancy of the allosteric site. Using the population-shift model (Eqs. 1 and 4) and the independently determined target and inhibitor affinities (Figure S1), we can simulate this allosteric behaviour (solid lines, bottom left and right) with high precision without the use of any floating parameters (see Figure S2 for details about correlation between estimated and experimentally observed values).