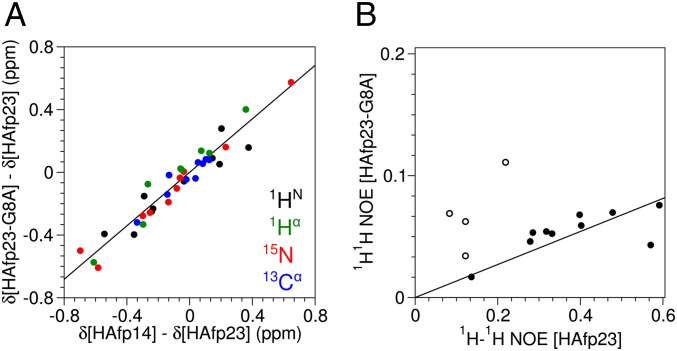
Fig. 5.
Population of the open conformations for HAfp23-G8A from (A) chemical shift and (B) 1H-1H NOE data. (A) The differences in the 1HN (black), 1Hα (green), 15N (red), and 13Cα (blue) backbone chemical shifts among HAfp23 (completely closed conformation), HAfp14 (completely open conformation), and HAfp23-G8A (open/closed mixture) at pH 7 show a high degree of correlation with a slope of 85 ± 4%, corresponding to the population of the open conformation(s) for HAfp23-G8A. The chemical shift differences have been scaled (δ*) relative to 1HN by the following factors derived from database SDs in secondary chemical shifts: 15N = 0.181, 13Cα = 0.315, 1Hα = 1.373. Chemical shifts from the site of mutation, residue 8, and the C-terminal residues of HAfp14, Gly13, and Trp14 were not included in the fit. (B) Intensities of interhelical NOEs in HAfp23 and HAfp23-G8A show a similar correlation with a slope of 14 ± 4%, corresponding to the population of the closed helical hairpin conformation. Open symbols correspond to NOEs between Met17 and the aromatic side chain of Phe9 and have been excluded from the fit, because they show evidence for close proximity in the open structures.