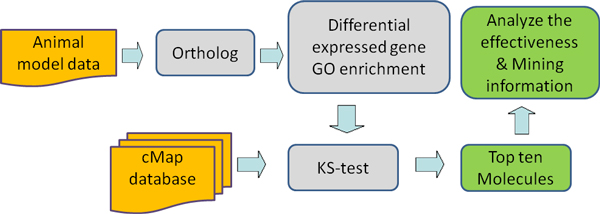
Figure 2.
Workflow of our analysis method. The cMap database of chemical interventions was used for search. Given an animal model query profile with gene id and expression value (treated/control), every animal gene id was converted to orthologous human gene id. Through differential expressed gene GO enrichment and KS-test with cMap database, a result was returned with a table of top 10 molecules which were most similar to that of the query profile, and their similar GO modules, positive or negative.