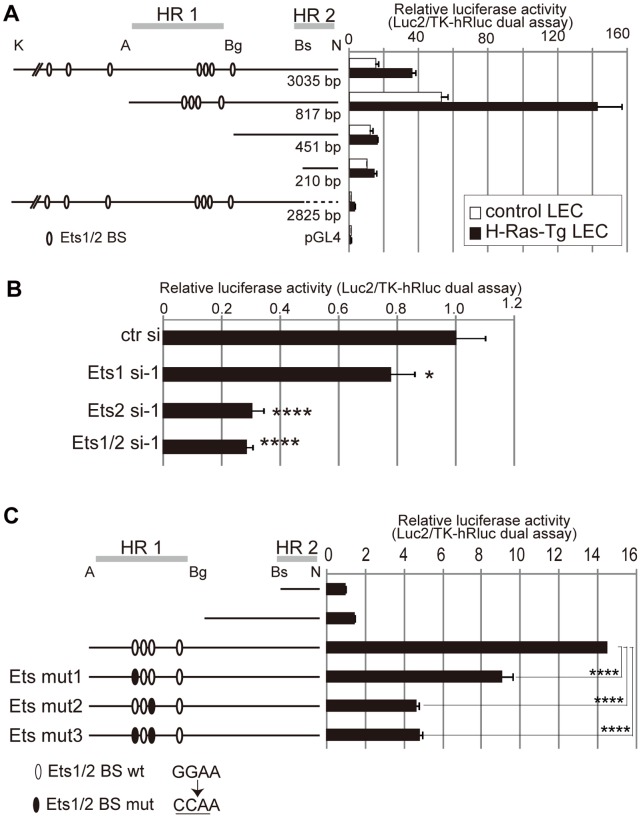
Figure 4. The 5′ regulatory region of the Vegfr3 gene drives luciferase expression in an Ets-dependent manner.
A. Luciferase assay using the pGL4-luc2 vector harboring variable lengths of the 5′ untranscribed and UTR regions of the mouse Vegfr3 gene in control and H-Ras-overexpressing mLECs. K, Kpn I; A, Avr II; Bg, Bgl II; Bs, BsiWI; N, Not I. A dashed line indicates the deletion between the BsiWI and Not I sites. Open ovals show putative Ets-binding sites predicted by the Transcription Element Search System (TESS; http://www.cbil.upenn.edu/tess). The previously reported homologous regions HR1 and HR2 [18] are also shown. Relative luciferase activities (luc2 vs. TK-hRluc vector-produced hRluc) are shown. Error bars represent the S.D.; n = 3. B. Luciferase assay using the pGL4-Luc2 vector harboring the Avr II-Not I fragment in control and Ets knockdown mLECs. Error bars represent the S.D.; n = 3. *p<0.05, ****p<0.001 (vs. mLECs transfected with control siRNA). C. Luciferase assay using the pGL4-Luc2 vector harboring the Avr II-Not I fragment with or without mutated Ets-putative binding sites (GGAA-to-CCAA mutation). Reporter vectors harboring the Bs-N fragment or Bg-N fragment were also assayed for comparison. Error bars represent the S.D.; n = 3. ****p<0.001 (vs. the wild-type Avr II-Not I fragment).