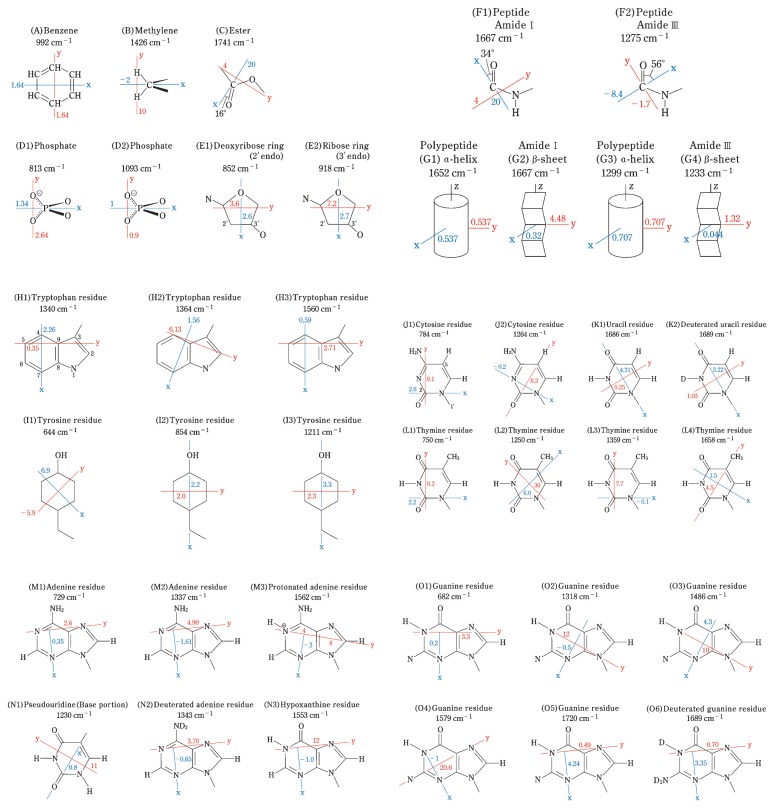
Abstract
The Raman scattering of a molecule is generated by interactions of its electrons with incident light. The electric vector of the Raman scattered light is related to the electric vector of the incident light through a characteristic Raman tensor. A unique Raman tensor exists for each Raman-active molecular vibrational mode. In the case of biologically important macromolecules Raman tensors have been determined for a few hundred vibrational Raman bands. These include proteins and their amino acid constituents, as well as nucleic acids (DNA and RNA) and their nucleotide constituents. In this review Raman tensors for 39 representative vibrational Raman bands of biological molecules are considered. We present details of the Raman tensor determinations and discuss their application in structural studies of filamentous bacteriophages (fd, Pf1, Pf3 and PH75), fowl feather rachis and eyespots of the protists, Chlamydomonas and Euglena.
Keywords: polarized Raman spectroscopy, Raman tensor, DNA, RNA, protein, filamentous bacteriophage
Introduction
Over the past six decades Raman spectra have been obtained on numerous molecules of biological interest. Surveys of applications to proteins and nucleic acids have been given recently.1),2) Typically, each Raman spectrum consists of about 50 discernible vibrational bands, to each of which a frequency in wavenumber units (cm−1) and a relative Raman scattering intensity (in arbitrary units) are designated. In some cases the spectral bands can be further characterized in terms of vibrational modes involving specific internal coordinates of the biomolecule by analogy with Raman spectra of simpler “model” compounds. Complete characterization of a given Raman band, however, requires that its Raman tensor also be determined. The Raman tensor associated with a Raman band describes how the polarizability of the molecule oscillates with the molecular normal mode of vibration. Methods for determining Raman tensors and for exploiting their use in structural studies of biomolecules and biomolecular complexes have been a major focus of our work in recent years.3)–6)
A Raman tensor is a 3 × 3 matrix that interrelates the electric vector (x1, y1, z1) of the exciting radiation with the electric vector (x2, y2, z2) of the Raman scattered radiation as given by Equation [1].
| [1] |
Because αxy = αyx, αyz = αzy and αzx = αxz, only six tensor components need be determined. Here, x, y, and z are rectangular coordinate axes that are fixed on the molecule but are otherwise chosen arbitrarily. If the principal axes of the Raman tensor, which are unique for any given Raman band, are selected as the xyz coordinate system, then the six non-zero tensor components are reduced to the three diagonal components, αxx, αyy and αzz. Thus, the Raman tensor can be established by determining αxx, αyy, and αzz, and three angular parameters that fix the tensor principal axes in the xyz coordinate system.
To define the principal axes system (xyz) of the local Raman tensor, we arbitrarily select three nonlinearly arranged atoms E1, E2 and A in the molecule of interest such that the y-axis is parallel to the line connecting atoms E1 and E2, the x-axis is parallel to the perpendicular line connecting atom A with the y-axis, and the z-axis is perpendicular to both the y and x axes. Although a set of selected axes may not encompass the entire 4π solid angle, it is generally sufficient for determining the Raman tensor. This follows from the fact that every interval between the directions of two candidate axes introduces an error that is small relative to the errors in experimental measurements.
Absolute values for the tensor components αxx, αyy and αzz are usually not required. The relative tensor magnitudes, which are defined by Equations [2a] and [2b], are sufficient:
| [2a] |
| [2b] |
Thus, the Raman tensor can be described in terms of the parameters A, E1, E2, r1 and r2.
Methods for Raman tensor determinations
The following three approaches have been used in combination with one another to determine Raman tensors of biological molecules. (i) Measurement of polarized Raman scattering on oriented single crystals of known molecular structure; (ii) measurement of Raman band depolarization ratios (ρ = I⊥/I||, where I⊥ and I|| are the Raman intensities of the perpendicular and parallel components of the Raman scattering) for molecules distributed randomly in solution; and (iii) ab initio molecular orbital calculations. We next consider each of these approaches.
1. Polarized Raman scattering of oriented single crystals
The crystal-fixed Raman tensor components (αaa, αbb, αcc, αbc, αab, αac) can be converted to the molecule-fixed components (αxx, αyy, αzz) through the following equations:
| [3a] |
| [3b] |
| [3c] |
| [3d] |
| [3e] |
| [3f] |
Here, lx, mx, . . . , ny, nz are the direction cosines of the x, y, and z axes in the rectangular abc-coordinate system. By the use of Equations [3a]–[3f], the polarized Raman intensity ratios can be given as:
| [4] |
| [5] |
where the notation Ijj signifies that the electric vectors of both the exciting and scattered radiation are along the j-axis (j = a, b, c).
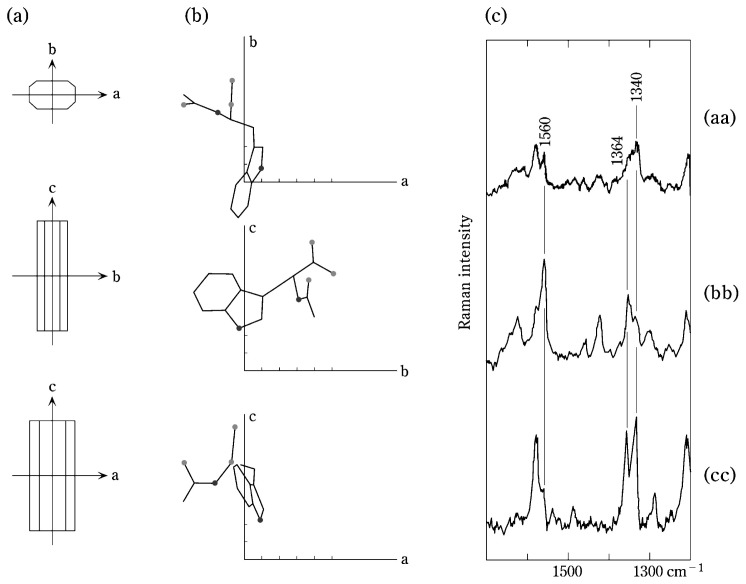
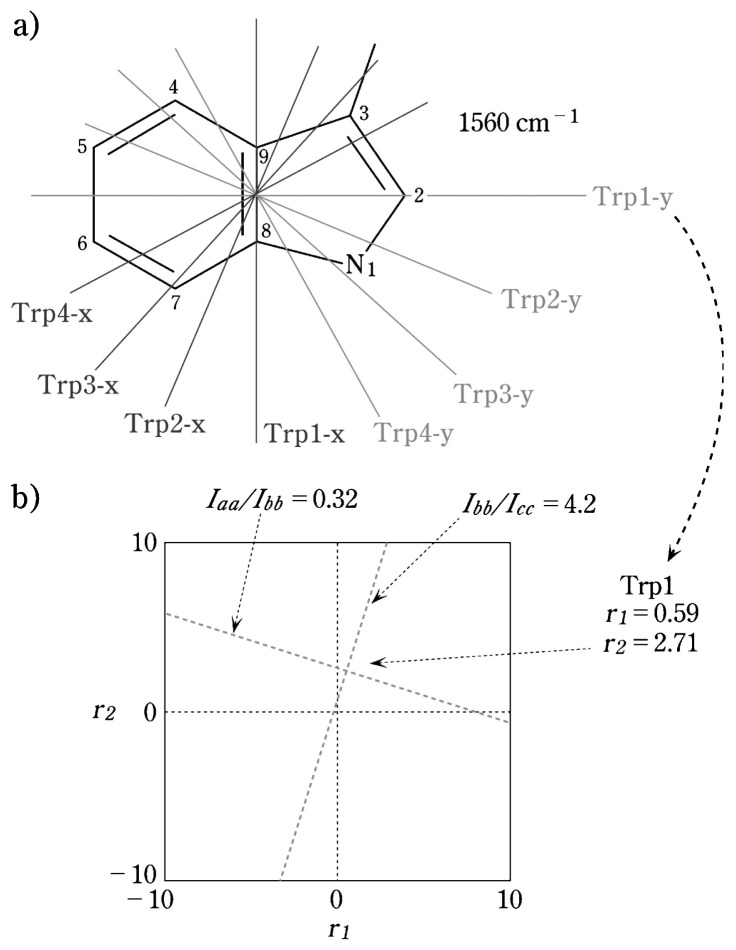
In the crystal lattice of N-acetyl-L-tryptophan7) (Fig. 1a), the molecules are oriented as shown in Fig. 1b. Polarized Raman spectra obtained on the crystal with 488.0 nm excitation are shown in Fig. 1c.8) To determine the Raman tensor of the 1560 cm−1 Raman band of tryptophan the following strategy was employed. Let us assume that the principal axes are those designated Trp1 in Fig. 2a, where E1 = C5, E2 = C9 and A = C7. This is one of four trial axis systems depicted in Fig. 2a and, as shown below, the most favorable for calculating the Raman tensor by method (ii), above. From the spectra of Fig. 1c, Iaa/Ibb = 0.32 and Ibb/Icc = 4.2. By the use of Equations [4] and [5], two intersecting contour lines can be drawn in r1/r2 space from which the tensor values r1 = 0.59 and r2 = 2.71 are obtained at the point of inter-section, as shown in Fig. 2b.8)
Fig. 1.
Orientation of tryptophan molecules in a single crystal of N-acetyl-L-tryptophan and their polarized Raman spectra. (a) crystallographic axes a, b and c in the single crystal of N-acetyl-L-tryptophan (orthorhombic, space group P2121217)). (b) Orientation of a tryptophan molecule in the unit cell. (Only one of the symmetrically equivalent molecules is shown.) (c) Polarized Raman spectra (488.0 nm excitation) in (aa), (bb) and (cc) orientations. In each orientation the incident and scattered electric vectors are parallel to one of the crystallographic axes, as defined above.8)
Fig. 2.
(a) Trial coordinate systems (Trp1, Trp2, Trp3, Trp4) that were assessed as principal axes (xyz) suitable for calculating the Raman tensors of tryptophan. (b) Calculated Raman intensity ratios in r1,r2-space. The contours are drawn for the Trp1 axis system in (a).
2. Depolarization ratios of randomly distributed molecules
The Raman depolarization ratio (ρ) is defined for randomly oriented molecules in solution as the ratio of the Raman scattering intensities polarized along directions perpendicular (I⊥) and parallel (I||) to that of the incident radiation. In terms of r1 and r2, ρ is given by Equation [6].
| [6] |
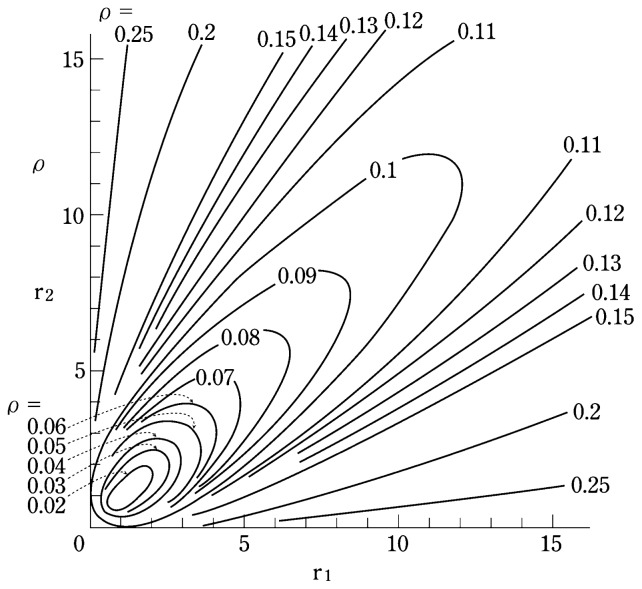
This relationship between the depolarization ratio (ρ) and the Raman tensor ratios (r1 and r2)9),10) is presented graphically in Fig. 3. Although Equation [6] does not provide information on the orientations of the tensor principal axes with respect to the molecular framework, it does provide a measure for judging the appropriateness of the principal axis system chosen in calculating the Raman tensor. For example, in determining the Raman tensor of the 1360 cm−1 tryptophan band, four principal axes systems, Trp1, Trp2, Trp3, and Trp4 (Fig. 2a) were evaluated. Using the experimentally determined values, Iaa/Ibb = 0.43 and Ibb/Icc = 0.58 (Fig. 2b), the following results were obtained: Trp1 (r1 = 2.85, r2 = 1.43, ρ = 0.06); Trp2 (r1 = 1.56, r2 = 6.13, ρ = 0.15); Trp3 (r1 = 2.20, r2 = 3.34, ρ = 0.05); and Trp4 (r1 = 2.04, r2 = 2.95, ρ = 0.04). The experimentally determined depolarization ratio is ρ = 0.17 ± 0.03, and therefore, only the Trp2 axis system is consistent with the experimental data. It is thus concluded that when 488.0 nm excitation is employed the Raman tensor of the 1360 cm−1 band of tryptophan is well represented by the parameters E1 = C4, E2 = C2, A = C7, r1 = 1.56, r2 = 6.13.8)
Fig. 3.
A contour diagram showing the relationship between the depolarization ratio (ρ) and the shape of the Raman tensor (r1, r2) of a randomly oriented molecule or molecular subgroup.9) The relationship is plotted as a contour map in the r1,r2-plane with values of r1 and r2 ranging from 0 to ~15. A similar map with the values of r1 and r2 in the range from −15 to +15 is available has been shown previously.4) Additional maps with expanded scales are also available.9)
3. Ab Initio molecular orbital calculation
With recent advances in computational capacity it is now feasible to employ a larger basis set in ab initio calculations, enabling improved calculation of the vibrational modes of a molecule. Ab initio self-consistent force field molecular orbital (MO) calculations were carried out on thymine using HONDO 8.5 as follows.11) First, geometry optimization and force constant calculations were done using the 6-31-G basis set. Second, normal modes of vibration were calculated using the 6-31-G basis set augmented with diffuse p- and d-type functions.11) Third, calculations were made of the derivatives of the molecular polarizability with respect to the atomic displacements along the Cartesian coordinate axes (chosen arbitrarily). Finally, using the calculated normal modes of vibration, the polarizability derivatives with respect to the normal coordinates were calculated.12) These are the Raman tensors calculated with the coordinate axis system chosen above.
Most of the intense bands observed in the Raman spectrum of thymine (488.0 nm excitation) are attributable to in-plane vibrations.11) The Raman tensor of an in-plane vibration can be easily calculated. The tensor has only four components, αxx, αyy, αxy and αzz and with the proper in-plane rotation of the axis system (into x′y′z) the off-diagonal component αxy can be eliminated. This rotation results in the principal axis system x′y′z and the relative magnitudes of the diagonal components α′xx, α′yy and α′zz being defined as r1 = α′xx/α′zz and r2 = α′yy/α′zz. This calculation also provides the depolarization ratio ρ for every band in the Raman spectrum of thymine.
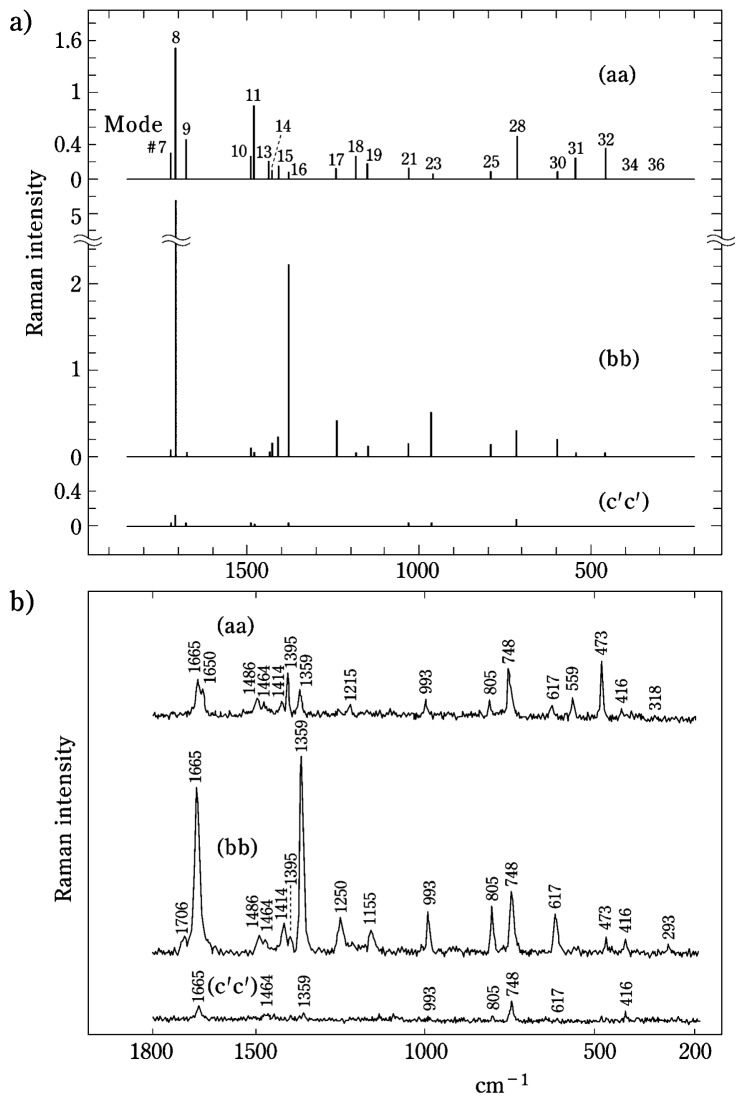
The Raman scattering anisotropies expected for a single crystal of thymine13) are calculated utilizing the thymine Raman tensors as described above and are compared with the experimentally determined values in Fig. 4. This comparison provides a test of both the validity of the “oriented gas” model of a crystal, as well as a test of the reliability of the tensors calculated by the ab initio method. Figure 4 reveals that the general features of the calculated and observed polarized Raman spectra (Figs. 4a and 4b, respectively) are very similar.
Fig. 4.
(a) Raman scattering anisotropies calculated for a single crystal of thymine on the basis of Raman tensors derived from ab initio MO calculations.11),12) The intensity scale is in units of Å2/amu−1/2. Top trace (aa): Raman frequencies and intensities expected when the electric vectors of the exciting and scattered beams are parallel to the crystallographic a axis. Middle trace (bb): Raman frequencies and intensities expected when the electric vectors of the exciting and scattered beams are parallel to the crystallographic b axis. Bottom trace (c′c′): Raman frequencies and intensities expected when the electric vectors of the exciting and scattered beams are parallel to the crystallographic c′-axis, where the c′-axis is defined as the axis that is perpendicular to the a axis in the ac-plane. (b) Polarized Raman spectra obtained on a single crystal of thymine (monoclinic, P21/c13)) with 488.0 nm excitation.12) The three spectra shown (aa, bb, c′c′) were obtained with orientations of the crystallographic axes with respect to the electric vectors of the exciting and scattered beams as described in (a) above.
In the aa spectrum (Fig. 4b) Raman bands (relative intensities) are observed at 993 (weak), 805 (weak), 748 (strong), 617 (weak), 559 (medium) and 473 cm−1 (strong). These bands and their relative intensities are satisfactorily reproduced in the calculated aa spectrum Fig. 4a). In the bb spectrum bands are observed at 1665 (strong), 1359 (strong), 1250 (medium), 1155 (weak), 993 (medium), 805 (medium), 748 (strong), 617 (medium), 473 (weak) and 416 cm−1 (weak) (Fig. 4b), in remarkably good agreement with the calculated bb spectrum (Fig. 4a). In the experimentally determined c′c′ spectrum only the two bands at 1665 and 748 cm−1 exhibit significant intensities, and again these are reproduced in the calculated c′c′ spectrum.
We note that the ab initio MO calculation on the 1241 cm−1 band of thymine provides the values r1 = 4.0 and r2 = 38 and ρ = 0.27.12) These results compare favorably with the experimental values (r1 = 6.0, r2 = 30, ρ = 0.28) determined from polarized Raman measurements on a thymine single crystal. The present findings support the view that Raman tensors calculated by the ab initio method as outlined above are significantly more reliable than in the past. The ab initio method represents a potentially powerful tool for use in the interpretation and analysis of vibrational spectra.
While the agreement between experimental and calculated Raman spectra of Figs. 4a and 4b is generally quite good, a number of minor discrepancies are apparent. The following are possible contributing factors: (i) The ab initio calculations are carried out on an isolated thymine molecule, whereas the experimental results are obtained for thymine in the crystal state, where O and N atoms are likely to be involved in strong hydrogen bonds. (ii) The calculation of the derivatives of the molecular polarizabilities assumes a static electric field, whereas the experiments are carried with an excitation wavelength (488.0 nm) that may introduce resonance effects. (iii) A small Raman tensor may have a large error associated with its determination if the crystal axes are tilted only slightly from their presumed correct orientations with respect to the directions of polarization of the exciting and scattered beams. A detailed examination of these factors may lead to further improvement and refinement of the ab inito methods for use in tensor determinations.
A catalog of Raman Tensors
By use of the methods outlined above Raman tensors have now been determined for approximately 200 Raman bands that originate from atomic group vibrations of biologically important amino acid and nucleotide building blocks of proteins and DNA. These tensors are listed in Table 1 with appropriate annotation. Of the tensors listed in Table 1, a representative selection of thirty-nine was selected for detailed illustration in Fig. 5. Unless indicated otherwise, all were obtained at off-resonance or pre-resonance conditions using either 488.0, 514.5 or 532.0 nm excitation. (Raman tensors determined at conditions of resonance excitation are discussed elsewhere.)
Table 1.
Raman tensors of biological moleculesa
| sample (state) | group | Raman cm−1 | reference |
|---|---|---|---|
| B DNA (fiber) | guanine | 682 | Thomas et al.10) |
| adenine | 729 | ||
| cytosine | 784 | ||
| thymine | 750, 1669 | ||
| 2′CH2 | 1420 | ||
| 5′CH2 | 1465 | ||
| A DNA (fiber) | guanine | 666 | Thomas et al.10) |
| adenine | 731 | ||
| cytosine | 784 | ||
| thymine | 753, 1667 | ||
| 2′CH2 | 1419 | ||
| 5′CH2 | 1463 | ||
| Z DNA (crystal) | guanine | 625, 670, 1318, 1486, 1579 | Benevides et al.16) |
| cytosine | 598, 748, 1264 | ||
| 2′CH2 | 1426 | ||
| 5′CH2 | 1433 | ||
| dsRNA (fiber) | guanine | 668, 724, 1377, 1482, 1720 | Benevides et al.44) |
| guanine·D2O | 664, 716, 1372, 1476, 1689 | ||
| adenine | 1337 | ||
| adenine·D2O | 1343 | ||
| cytosine | 598, 785, 1250 | ||
| cytosine·D2O | 571, 777, 1255 | ||
| uracil | 1686 | ||
| UpA (crystal) | adenine | 1562 | Tsuboi et al.45) |
| adenine·H+ | 713, 1332, 1516, 1560 | Ushizawa et al.46) | |
| uracil | 790, 1238, 1626, 1672, 1696 | ||
| PO2− | 1080 | ||
| P–O | 801 | ||
| cytidine (crystal) | cytosine | 785, 1264 | Tsuboi et al.47) |
| cytosine | 603, 793, 1254, 1536 | Ueda et al.48) | |
| 5′CH2 | 1437 | Tsuboi et al.47) | |
| thymidine (crystal) | thymine | 494, 632, 675, 771, 793, 1017, 1066, 1235, 1365, 1642, 1665 | Ushizawa et al.14) |
| 2′CH2 | 1404 | Tsuboi et al.47) | |
| 5′CH2 | 1403 | Ushizawa et al.14) | |
| thymine (crystal) | thymine | 456, 545, 596, 714, 793, 1027, 1241, 1379, 1710 | Tsuboi et al.12) |
| AZT (crystal) | thymine | 495, 771, 1238, 1355, 1392, 1644, 1658, 1666, 1690 | Kumakura et al.49) |
| 5-iodouracil (crystal) | 5-iodouracil | 600, 624, 790, 1325, 1610, 1645 | Ueda et al.50) |
| Ψ-uridine (crystal) | α-pseudouracil | 553, 635, 789, 1000, 1150, 1230, 1340, 1368, 1417, 1490, 1654, 1666 | Ueda et al.51) |
| ATP (crystal) | adenine | 729, 1413 | Tsuboi et al.52) |
| adenine | 724, 1125, 1324, 1405, 1501, 1553, 1604 | Ueda et al.53) | |
| adenine (crystal) | adenine·2H+ | 722, 1320, 1424, 1585 | Ushizawa et al.54) |
| IMP (crystal) | hypoxanthine | 730, 899, 1381, 1485, 1553, 1594, 1678 | Ushizawa et al.55) |
| 5′CH2 | 1425 | Ushizawa et al.55) | |
| silk & feather | amide I (β) | 1665 | Tsuboi et al.56) |
| amide I (β) | 1664b | Tsuboi et al.57) | |
| amide III (β) | 1233 | Yokote et al.41) | |
| silk & feather | amide III (α) | 1299 | Yokote et al.41) |
| aspartame (crystal) | amide I | 1667 | Tsuboi et al.3) |
| amide III | 1275 | Tsuboi et al.47) | |
| ester C=O | 1741 | ||
| phenylalanine | 1204 | ||
| fd (fiber) | amide I (α) | 1651 | Overman et al.25) |
| Pf1 (fiber) | CαH (α) | 1328, 1340 | Tsuboi et al.58) |
| tyrosine (crystal) | tyrosine | 432, 642, 829, 1179, 1200, 1614 | Tsuboi et al.15) |
| CO2− | 1327 | Tsuboi et al.15) | |
| CH2 | 1434 | Tsuboi et al.15) | |
| tyrosine-2,3,5,6-d4 (crystal) | |||
| CD | 2273, 2290 | Tsuboi et al.38) | |
| N-acetyl-tryptophan (crystal) | |||
| tryptophan | 757, 1010, 1332, 1357, 1424, 1458, 1487, 1557, 1576, 1617, 3416 | Tsuboi et al.8) | |
| benzene (crystal) | ring | 992 | Ueda et al.9) |
| ring | 1600 | Tsuboi et al.47) | |
The table provides a comprehensive summary of biological molecule Raman tensors from presently available publications cited in the right-hand column. Column 1 lists the class of biological compound (sample state where relevant is given in parentheses); column 2 identifies the molecular group or subgroup; column 3 is the Raman wavenumber. All data were obtained from spectra excited at 488.0, 512.5 or 532 nm, unless indicated otherwise.
Raman spectrum excited at 785 nm.
Fig. 5.
Raman tensors determined for selected Raman bands (488.0, 514.5 and/or 632.0 nm excitation) of several nucleic acid and protein substituents or subgroups. For each tensor, the Raman frequency (cm−1), principal axes, and tensor components (r1 and r2) are given. The principal axes are drawn in blue (x axis) and red (y axis). The z axis is perpendicular to the (xy) plane. The tensor components r1 = αxx/αzz and r2 = αyy/αzz are denoted by blue and red numerals, respectively. References to the original literature, where details of the tensor calculations are described, are as follows: (A) Ueda et al.;9) (B) Benevides et al.;16) (C) Tsuboi et al.;3),47) (D1) Benevides et al.;44) (D2) Thomas et al.;10) (E1) Ushizawa et al.;14) (E2) Benevides et al.;44) (F1) Tsuboi et al.;3) (F2) Tsuboi et al.;3) (G1) Overman et al.;25) (G2) Tsuboi et al.;57) (G3) Yokote et al.;41) (G4) Yokote et al.;41) (H1)(H2)(H3) Tsuboi et al.;8) (I1)(I2)(I3) Tsuboi et al.;15) (J1) Thomas et al.;10) (J2) Benevides et al.;16) (K1)(K2) Benevides et al.;44) (L1) Thomas et al.;10) (L2)(L3) Tsuboi et al.;12) (L4) Kumakura et al.;49) (M1) Thomas et al.;10) (M2) Benevides et al.;44) (M3) Ushizawa et al.;46) (N1) Ueda et al.;51) (N2) Benevides et al.;44) (N3) Ushizawa et al.;55) (O1) Thomas et al.;10) (O2)(O3)(O4) Benevides et al.;16) (O5)(O6) Benevides et al.44)
Transferability of Raman tensors
As shown in Fig. 5 (L2), the Raman tensor of the 1250 cm−1 band of thymine is defined by r1 = 6.0, r2 = 30, with the y-axis aligned approximately along the C4—N1 line. This tensor was determined by polarized Raman spectroscopy of a thymine monoclinic single crystal (space group P21/c).12) For comparison, the Raman tensor of the 1235 cm−1 band of thymidine has r1 = 4.3 and r2 = 20.1, with the y-axis positioned along the C4—N1 line. In this case the tensor was determined from polarized Raman spectra of a thymidine single crystal in space group P212121.14) Thus, the replacement of the thymine N1 proton by the deoxyribosyl moiety does not greatly impact the shape of this particular Raman tensor. In general, however, transferability of Raman tensors from one molecular derivative to another should not necessarily be presumed. Nevertheless, in the analysis of molecular structures by polarized Raman spectroscopy, the transferability of the Raman tensor from an atomic group in a simple molecule to that in a macromolecular assembly like a virus, is often assumed. As a means of assuring that tensor transferability is not an issue, it is preferable to carry out structure analyses using several tensors for the same atomic group. For example, in determining the orientation of a tryptophan side chain in a viral protein, the use of Raman tensors for the 1557, 1357 and 1332 cm−1 Raman bands of tryptophan is recommended.8) For the tyrosine side chain, the Raman tensors for tyrosine bands at 1200, 829 and 642 cm−1 are often employed in combination with one another.15)
Raman tensors in structural studies of living systems
In determining molecular orientations in a biological assembly the goal is to determine the Eulerian angles of the constituent molecules (or molecular subgroups) using polarized Raman spectral measurements and known Raman tensors. The set of equations required to determine Eulerian angles are given above [Equation [3]]. In the sample coordinate system (abc), the orientation of the atomic group (xyz) is given by direction cosines (lx, · · ·, nz) through the Raman tensor (αxx, αyy, αzz).16),17) Several applications are next discussed.
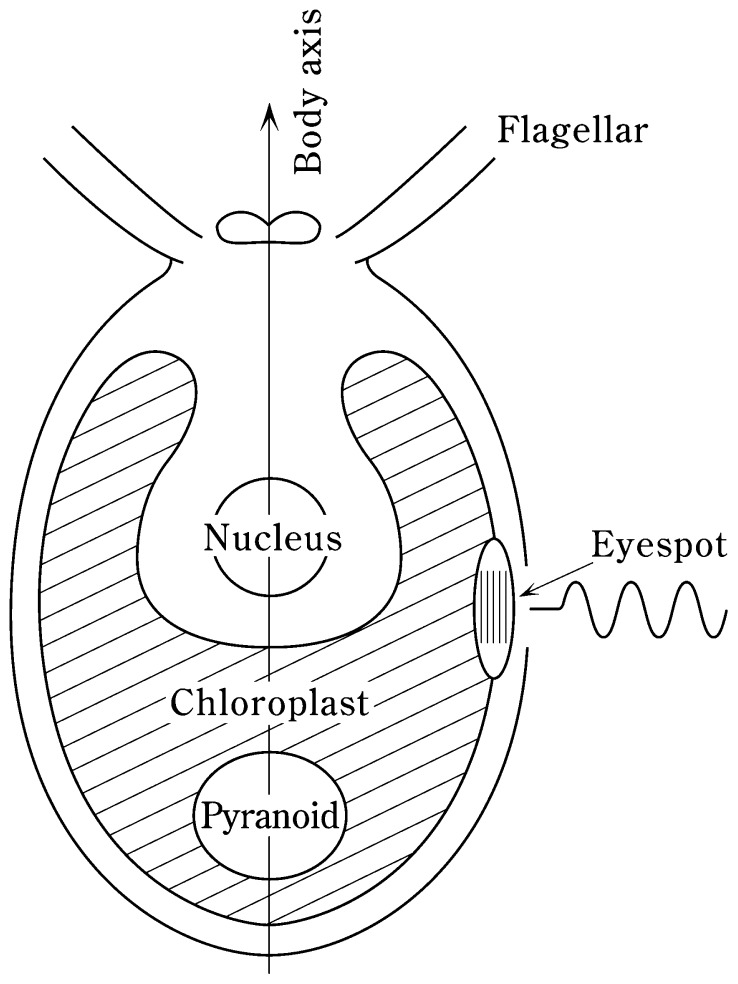
1. Carotenoids in protists
Single cells of Euglena and Chlamydomonas were immobilized and their polarized Raman scattering spectra (514.5 nm excitation) were mapped at several points with spatial resolution of approximately 1 μm.17) At each point intense Raman bands of carotenoid pigments were observed at 1530 and 1159 cm−1. The eyespots of both protists exhibited relatively high carotenoid contents, while smaller amounts of carotenoid were uniformly distributed throughout the chloroplasts. The Chlamydomonas eyespot exhibited an elongated shape with approximate dimensions of 1 × 2 μm. Polarization analysis revealed that the carotenoid chains within the Chlamydomonas eyespot are aligned preferably along the long axis of the eye-spot, which is itself parallel to the long axis of the body of the alga (Fig. 6).17),18) Thus, it appears that Chlamydomonas detects light polarized parallel to the long axis of its body. In contrast to Chlamydomonas, the eyespot of Euglena does not exhibit anisotropy in polarized light detection.17)
Fig. 6.
Location and orientation of the Chlamydomonas eye-spot. Carotenoid molecules are aligned along the long axis of the eyespot, which in turn is parallel to the long axis of the body of the alga.
2. Filamentous bacteriophages
In filamentous bacteriophages the constituent molecules exhibit symmetry with respect to the filament axis (c). Molecular axial symmetry is also encountered in fowl feather barb. In such axially symmetric molecular assemblies it is a simple matter to prepare for polarized Raman measurements an oriented fiber containing the filamentous bacteriophage (or feather barb) in which the assemblies are oriented unidirectionally along c. The a and b axes, which are perpendicular to c, are equivalent to one another. For such oriented fibers only the polarized Raman intensities Ibb, Icc, Ibc and Iab need be measured. The tensor coordinates (xyz) of each Raman band can be located with respect to the coordinates of the fiber sample (abc) by determining two parameters, the Eulerian angles θ and χ. Here, θ is the angle by which the tensor z axis is tilted from the fiber axis (c) and χ is the angle between the tensor y axis and the line of intersection (O–N) of the xy and ab planes. The polarized Raman intensity ratios Icc/Ibb and Icc/Ibc are given in terms of θ and χ by Equations [7] and [8]:
| [7] |
| [8] |
Orientation of DNA in bacteriophage Pf1
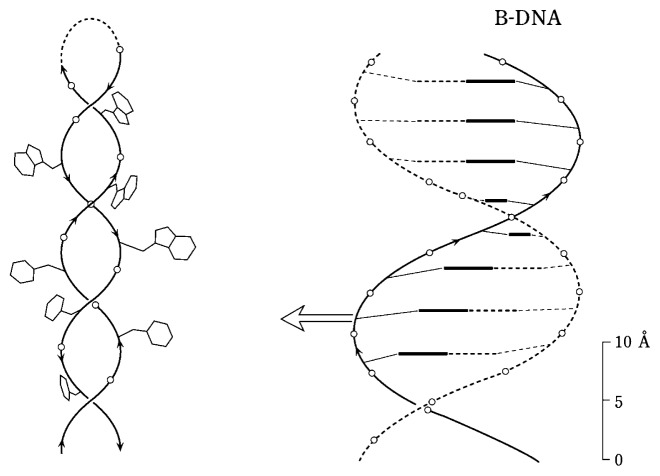
Bacteriophage Pf1 (~6.5 × 2000 nm filament) comprises a covalently closed single-stranded DNA genome of 7349 nucleotides coated by 7350 copies of a 46-residue α-helical subunit. X-ray fiber diffraction studies of Pf119) have been interpreted to signify that the DNA phosphates are located at a radius of 2.5 Å from the c axis and separated axially by a rise and rotation per nucleotide of 6.1 Å and 132°, respectively. Assuming this model we have exploited polarized Raman spectroscopy of Pf1 to determine the orientations of the DNA bases with respect to the c axis.20),21)
Polarized Raman intensities Icc, Ibb, Ibc and Icb were measured for the Raman bands of the DNA bases at 680 (dG), 720 (dA), 750 (dT) and 782 cm−1 (dC). By use of the known Raman tensors [Figure 5 (O1), (M1), (L1) and (J1)], the following angles of inclination (θ, χ) were determined for the base residues: dG (63°, 46°), dA (63°, 47°), dT (68°, 44°) and dC (67°, 46°). These results have been used to generate a model for the structure of Pf1 DNA within the viral capsid. The model is compared with the structure of canonical B DNA in Fig. 7. This new DNA model has a structure reminiscent of that proposed previously for double-helical DNA subjected to severe stress in the axial direction (termed P DNA and in which the bases extend outward from a centralized sugar-phosphate backbone).22) While P DNA is produced artificially by stretching B DNA and thus retains base complementarity in apposing strands, the Pf1 model of Fig. 7 is proposed to occur in vivo and irrespective of base composition.
Fig. 7.
(Left) Molecular model of DNA in bacteriophage Pf1.21) Base residues are shown extending outward from the phos-phodiester mainchain. (Right) The structure of canonical B DNA.
Orientation of coat protein α-helices in filamentous bacteriophages
The class I filamentous bacteriophages (fd, M13, f1) are important model systems for membrane protein assembly and are used extensively as cloning vectors and vehicles for peptide display.23) The virion (~6.5 × 880 nm) comprises a single-stranded DNA genome sheathed by ~2700 copies of a 50-residue α-helical subunit (pVIII).24) The average angle of inclination of the pVIII α-helix from the virion axis (c), as determined by polarized Raman analysis of the amide I band using the Raman tensor of Fig. 5 (G1), is 16 ± 4°.25) Similarly, tilt angles of 16 ± 4° were determined for class II phages Pf120) and Pf3.26) Interestingly, in the thermophilic bacteriophage PH75 a tilt angle of 25 ± 5° was determined.27)
Orientation of coat protein tryptophan residues in filamentous bacteriophages
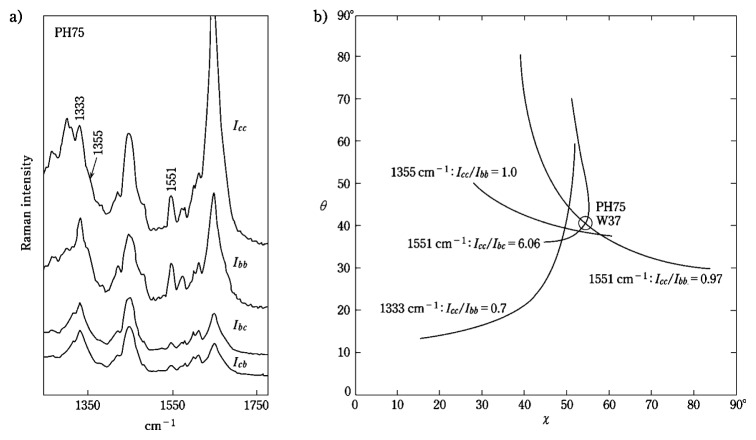
The coat protein subunit of bacteriophage PH75 has a single tryptophan residue (Trp 37),28)–30) which exhibits a prominent Raman marker at 1551 cm−1. Polarized Raman measurements on the 1551 cm−1 band in oriented fibers of PH75 indicate Icc/Ibb = 0.97 and Icc/Ibc = 6.06 (Fig. 8a).27) With Equations [7] and [8] and the Raman tensor of Fig. 5 (H3), one obtains the contour lines in θ,χ-space (Fig. 8b) that specify the coordinates of the Trp 37 indole ring orientation (θ = 41°, χ = 55°). Similar analyses of the Trp 38 markers at 1333 and 1355 cm−1 (Fig. 8b) in the polarized Raman spectra of PH75 are consistent with this indole ring orientation.
Fig. 8.
(a) Polarized Icc, Ibb, Ibc, and Icb Raman spectra (1300–1750 cm−1) obtained from an oriented fiber of PH75 that was excited at 532 nm. (b) Contour maps in θ,χ-space showing Icc/Ibb values for the 1333, 1355 and 1551 cm−1 Raman bands of Trp 37. The contour line is also shown for the ratio Icc/Ibc of the 1551 cm−1 band. On the basis of these results the Eulerian angles (θ, χ) that define the orientation of the Trp 37 indolyl plane with respect to the virion axis are: θ = 45° and χ = 59° (denoted by the open circle at the center of the plot).
A single tryptophan residue is also found in the coat subunit of bacteriophage fd (Trp 26)31),32) and in that of bacteriophage Pf3 (Trp 38).32) Polarized Raman spectroscopy of fd has yielded θ = 74°, χ = 61° for Trp 26 (Fig. 8a),33) while the corresponding analysis of Pf3 has yielded θ = 77°, χ = 45° for Trp 38 (Fig. 8b).26),27) These results refine and correct previously proposed models.24),34)–36)
Orientation of coat protein tyrosine residues in filamentous bacteriophages
The coat protein subunit of bacteriophage fd contains two tyrosines (Tyr 21 and Tyr 24),32),37) the side chain orientations of which have been determined independently by constructing single site mutants (Y21M and Y24M) and performing polarized Raman spectroscopic analyses of oriented fibers of each mutant virion.38) In addition, a stereo-array isotope labeling (SAIL) method was adopted, in which the virus was grown in the presence of L-tyrosine-2,3,5,6-d4 to incorporate deuterio-phenoxyl moieties into the coat protein subunits. The isolated Raman markers of the tyrosyl C–D stretching modes (Table 1) were then analyzed by polarized Raman spectroscopy in conjunction with the appropriately determined Raman tensors (Fig. 5 (I1), (I2) and (I3)].38) By the use of these methods the phenoxyl ring orientations of both Tyr 21 and Tyr 24 were found to be defined by θ = 71 ± 5°, χ = 36 ± 5°) (Fig. 9a).
Fig. 9.
Orientations of tryptophan and tyrosine residues in the protein subunits of bacteriophages: fd, Pf3 and PH75. The Trp 26 orientation as initially proposed in the model of Marvin et al.24) is represented by dotted lines in the fd illustration. The orientation determined experimentally by polarized Raman spectroscopy is also shown (solid lines).33)
The application of parallel methods to the class II filamentous bacteriophage Pf1 has indicated that the two tyrosines (Tyr 25 and Tyr 40) of its coat protein subunit are oriented with θ = 76 ± 10° and χ = 34 ± 10°, which is indistinguishable from those of the class I filamentous bacteriophage fd.20) This close correspondence between the subunit tyrosine orientations in class II (symmetry C1S5.4) and class I (C2S5) capsids suggests a conformational preference for α-helix packing irrespective of capsid symmetry. The two tyrosines (Tyr 15 and Tyr 39) of the coat protein subunit of the class II filamentous bacteriophage PH75, on the other hand, were found to depart from this pattern (θ = 80 ± 5°, χ = 33 ± 5°).27) This may be due to specific thermophilic requirements of phage PH75 or to the location of the subunit tyrosines in non-helical segments of the subunit.
Raman tensors in structural studies of biomaterials
Silk and feather keratin
The polypeptide chains of silks and feather keratins are composed primarily of antiparallel β-sheets arranged with axial symmetry.39) Raman tensors of the peptide group determined from Raman amide I and amide III bands are shown in Figs. 2 (F1) and 2 (F2), respectively. Using these local tensors and the atomic coordinates of the antiparallel β-sheet,40) the nonlocal amide I and amide III Raman tensors of Figs. 5 (G2) (r1 = 0.32, r2 = 4.48) and 5 (G4) (r1 = −0.044, r2 = 1.32) were determined. For both tensors the principal axes were defined with the z axis directed along the polypeptide chain in the plane of the sheet; the y axis was in the same plane perpendicular to z and approximately parallel to the peptide carbonyls; and the x axis was perpendicular to the plane of the β-sheet [yz plane, Figs. 5 (G2) and (G4)].
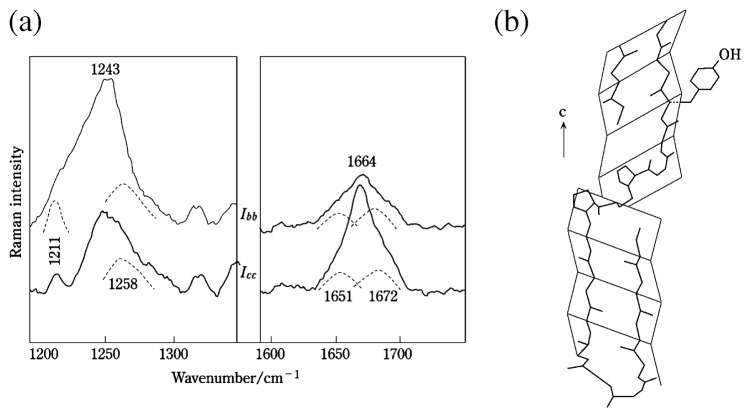
Silk and feather keratins may also contain varying amounts of β-turns that are expected to generate distinct Raman bands in the amide I (~1651 cm−1) and amide III (1258 cm−1) spectral regions. Therefore, the β-turn contributions should be accounted for when analyzing the spectrum for the β-sheet content.
Raman spectra of fowl feather rachis (Fig. 10) contain complex amide I and amide III features.41) In addition to the main peaks at 1664 and 1243 cm−1, satellite bands are observed at 1672, 1651 and 1258 cm−1. The satellite bands are assignable to unordered polypeptide chains and β-turns. They are presumed to contribute roughly equally to the Icc and Ibb polarized Raman spectra. Curve fitting of the Raman amide I and amide III bands was carried out with the assumption that Icc/Ibb = 0.28 for the band at 1664 cm−1 (amide I of 100% β-sheet), Icc/Ibb = 2.45 for the band at 1243 cm−1 (amide III of 100% β-sheet) and Icc/Ibb = 1, for the bands at 1672 and 1651 cm−1 (amide I of 0% β-sheet) and 1258 cm−1 (amide III of 0% β-sheet). The results indicate that 65 ± 10% of the feather rachis chains are nearly perfect β-sheets and 35 ± 10% are unordered polypeptide chains including β-turns. From amino acid sequence analysis a model of the polypeptide chain in the fowl feather rachis has been proposed.42) It consists of relatively short polypeptide chains (such as B-4, that approximates a rectangular block of 9.7 × 4.7 × 4.7 Å) connected through chains containing proline residues. Such a model, redrawn in Fig. 10, is consistent with spectral curve fitting results.
Fig. 10.
(a) Polarized Raman spectra of fowl feather rachis in the 1150–1300 cm−1 and 1600–1750 cm−1 spectral regions.41) (b) Structures and orientations of atomic groups in fowl feather rachis and barb that are consistent with the polarized Raman experimental results.41),42)
Polarized Raman studies have also been reported for oriented specimens of the fibroin fiber of B. mori silk, the spider dragline of N. edulis, and the fibroin silk thread of S. c. ricibi.43) In each case, a strong and relatively sharp amide I band with Icc/Ibb = ~0.30 occurs at 1667 cm−1 with no satellite band apparent at 1651 cm−1. This indicates that these fibers are deficient in unordered polypeptide chains and consist of mostly long pleated sheets having very little β-turn.
Conclusions
An extensive catalog of Raman tensors is now available for the further application of polarized Raman spectroscopy (488–532 nm excitation range) in structural biology. Because the Raman technique is non-invasive, the available tensors should prove particularly useful for in situ structural characterizations of living systems. Extension of laser excitation wavelengths to the near-UV and near-IR is expected to expand the use of polarized Raman spectroscopy dramatically, in particular to include intensely fluorescent molecules that are not amenable to study using excitation wavelengths in the visible range. Also worthy of further examination is the relationship between Raman tensor values and the nature of molecular vibrational modes. By use of ab initio MO calculations (briefly reviewed here), a better understanding of molecular electronic structures is anticipated.
Acknowledgments
The support of the Japan Ministry of Education, Science and Culture and the U.S. National Institutes of Health (Grants GM54378 and GM50776 to GJT) is gratefully acknowledged.
Abbreviations
- AZT
azidothymidine
- ds
double-stranded
- sc
single crystal
- UpA
uridylyl-(3′,5′)-adenosine monophosphate
- ATP
adenosine triphosphate
- IMP
inosine monophosphate
- SAIL
stereo-array isotope labeling
Biographies
Profile
Professor Masamichi Tsuboi was born in 1925, and started his research career in 1947 under the supervision of Professor San-ichiro Mizushima at Chemical Institute, University of Tokyo. He developed polarized infrared spectroscopy and applied it to studies of some polypeptides, the PO2− group of DNA, and OH groups in cellulose. This became the subject of the “Award for Young Chemist” which was given to him from the Chemical Society of Japan in 1958. In 1961, he was picked up by Faculty of Pharmaceutical Sciences, University of Tokyo, as the first Professor of physical chemistry in that Faculty. For 25 years since this time, he worked on the molecular structures of nucleic acids, proteins, and related drugs. Some parts of his such studies are summarized in his book “Drug-DNA Interactions—Structures and Spectra” (with co-authorship of Professor Kazuo Nakamoto and Dr. Gary D. Strahan, John Wiley 2008). For his work on Rice Dwarf Virus RNA (in collaboration with Professor Kin-ichiro Miura and Professor Yoichi Iitaka), Matsunaga Prize was awarded, and for resonance Raman studies of transfer RNAs (in collaboration with Dr. Susumu Nishimura) Chemical Society Japan Award. After his retirement from the University of Tokyo (1986), he started polarized Raman microscopy of biological systems in situ, in Iwaki-Meisei University. His work has been supported, not only by University of Tokyo (1947–1986), but also by a number of short term visiting professorships, in Johns Hopkins University (1964–1965), Pennsylvania State University (1969), Institute for Molecular Science (Okazaki, 1979–1981), Meisei University (1986–1987), Marquette University (Milwaukee, 1987–1989), and University of Missouri (Kansas City, 1990–2008). He is a member of Steering Committee, International Conference of Raman Spectroscopy, and in 1986–1988 he was its Chairman. He was awarded the Japan Academy Prize in 1987.

Profile
James Benevides was born in Fall River, Massachusetts in 1952. He completed undergraduate studies in zoology at the University of Massachusetts at Amherst (USA) and Masters and Ph.D. studies in the biological sciences at the University of Rhode Island (USA). This was followed by postdoctoral studies in molecular biophysics with Professor George Thomas at the University of Massachusetts at North Dartmouth. It was during this time that he became interested in the application of vibrational spectroscopy for structural studies on DNA and proteins. During the early 80’s, he along with Professor Thomas and collaborators at Massachusetts Institute of Technology initiated laser Raman studies on single crystals of DNA. They were the first to demonstrate that crystal packing forces could significantly impact the structure of B-form DNA. They also demonstrated that RNA-DNA hybrid structures were not always dominated by the RNA component. In the mid 80’s he had the opportunity to visit with Professor Masmichi Tsuboi at the University of Tokyo. This was the beginning of a long friendship and a fruitful collaboration with the Thomas Laboratory that led to the development of polarized Raman spectroscopy as a tool for structural studies on DNA and viruses. For the last twenty years Dr. Benevides has been on the faculty at the University of Missouri-Kansas City in the School of Biological Sciences where, in addition to polarized Raman studies, he has utilized Raman spectroscopy to investigate DNA-protein interactions in gene regulatory complexes and viruses.

Profile
Professor George J. Thomas, Jr. was born in New Bedford, Massachusetts on 24 December 1941. He earned his B.S. in chemistry (summa cum laude) at Boston College in 1963, and his Ph.D. in biophysical chemistry at the Massachusetts Institute of Technology (M.I.T.) in 1967. His dissertation research, which was conducted under the supervision of Professor Richard C. Lord, involved the development of Raman spectroscopy as a structural probe of DNA and RNA. He conducted postdoctoral research on nucleic acid structure with Nobel Laureate M. H. F. Wilkins at King’s College (London) in 1967–1968. During the years 1968–1987 Professor Thomas was a member of the Chemistry faculty at the University of Massachusetts in Dartmouth, where he served also as Department Chair. He has also held visiting scientist appointments at the Institute for Protein Research, Osaka University (1975–1976) and Department of Biology, M.I.T. (1982–1983). At the University of Massachusetts he equipped the first state-of-the-art Raman spectroscopy facility for research on biomolecular structures, and directed his research focus toward elucidating structures of nucleoprotein assemblies, including viruses. His landmark studies on viruses have demonstrated the unique advantages of Raman spectroscopy as a probe of protein-nucleic acid recognition. Since 1987 Professor Thomas has served as Distinguished Curators’ Professor and Head of Cell Biology and Biophysics in the School of Biological Sciences at the University of Missouri-Kansas City. His research, which is currently centered on molecular mechanisms of virus assembly, has been funded continuously since 1970 by grants from the U.S. National Institutes of Health and National Science Foundation. He was elected a Fellow of the American Association for the Advancement of Science in 2000 and has held appointments to N.I.H. study sections, N.S.F. review panels, various journal editorial boards and organizing committees of national and international conferences. He has also consulted for a number of pharmaceutical companies.

References
- 1).Benevides J. M., Overman S. A., Thomas G. J., Jr (2005) Raman, polarized Raman and ultraviolet resonance Raman spectroscopy of nucleic acids and their complexes. J. Raman Spectrosc. 36, 279–299 [Google Scholar]
- 2).Benevides J. M., Overman S. A., Thomas G. J., Jr (2004) Raman spectroscopy of proteins. InCurrent Protocols in Protein Science (eds. Coligan J. E., Dunn B. M., Ploegh H. L., Speicher D. W., Wingfield P. T.). John Wiley & Sons, New York, pp 17.8.1–17.8.35 [DOI] [PubMed] [Google Scholar]
- 3).Tsuboi M., Ikeda T., Ueda T. (1991) Raman microscopy of a small uniaxial crystal: Tetragonal aspartame. J. Raman Spectrosc. 22, 619–626 [Google Scholar]
- 4).Tsuboi M., Thomas G. J., Jr (1997) Raman scattering tensors in biological molecules and their assemblies. Appl. Spectrosc. Revs. 32, 263–299 [Google Scholar]
- 5).Tsuboi M. (2002) Raman scattering anisotropy of biological systems. J. Biomed. Opt. 7, 435–441 [DOI] [PubMed] [Google Scholar]
- 6).Tsuboi M., Thomas G. J., Jr (2007) Polarized Raman and polarized infrared spectroscopy of proteins and protein assemblies. InProtein Structures: Methods in Protein Structure and Stability Analysis (eds. Uversky V. N., Permyakov E. A.). Nova Science Publishers, Hauppage, New York, pp 153–194 [Google Scholar]
- 7).Yamane T., Andou T., Ashida T. (1977) N-acetyl-l-tryptophan. Acta Crystallog. B 33, 1650–1653 [Google Scholar]
- 8).Tsuboi M., Ueda T., Ushizawa K., Ezaki Y., Overman S. A., Thomas G. J., Jr (1996) Raman tensors for the tryptophan side chain in proteins determined by polarized Raman micro-spectroscopy of oriented N-acetyl-L-tryptophan crystals. J. Mol. Struct. 379, 43–50 [Google Scholar]
- 9).Ueda T., Ushizawa K., Tsuboi M. (1993) Depolarization of Raman scattering from some nucleotides of RNA and DNA. Biopolymers 33, 1791–1802 [DOI] [PubMed] [Google Scholar]
- 10).Thomas G. J., Jr, Benevides J. M., Overman S. A., Ueda T., Ushizawa K., Tsuboi M., et al. (1995) Polarized Raman spectra of oriented fibers of A DNA and B DNA: anisotropic and isotropic local Raman tensors of base and backbone vibrations. Biophys. J. 68, 1073–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11).Aida M., Kaneko M., Dupuis M., Ueda T., Ushizawa K., Tsuboi M., et al. (1997) Vibrational modes in thymine molecule from an ab initio MO calculation. Spectrochim. Acta A 53, 393–407 [Google Scholar]
- 12).Tsuboi M., Kumakura A., Aida M., Kaneko M., Dupuis M., Ushizawa K., et al. (1997) Raman scattering tensors in thymine molecule from an ab initio MO calculation. Spectrochim. Acta A 53, 409–419 [Google Scholar]
- 13).Ozeki K., Sakabe N., Tanaka J. (1969) The crystal structure of thymine. Acta Crystallog. B 25, 1038–1045 [Google Scholar]
- 14).Ushizawa K., Ueda T., Tsuboi M. (1997) Raman scattering tensors of thymidine. J. Mol. Struct. 412, 169–179 [Google Scholar]
- 15).Tsuboi M., Ezaki Y., Aida M., Suzuki M., Yimit A., Ushizawa K., et al. (1998) Raman scattering tensors of tyrosine. Biospectroscopy 4, 61–71 [DOI] [PubMed] [Google Scholar]
- 16).Benevides J. M., Tsuboi M., Wang A. H. J., Thomas G. J., Jr (1993) Local Raman tensors of double-helical DNA in the crystal: A basis for determining DNA residue orientations. J. Am. Chem. Soc. 115, 5351–5359 [Google Scholar]
- 17).Kubo Y., Ikeda T., Yang S.-Y., Tsuboi M. (2000) Orientation of carotenoid molecules in the eyespot of alga: In situ polarized resonance Raman spectroscopy. Appl. Spectrosc. 54, 1114–1119 [Google Scholar]
- 18).Yang S. Y., Tsuboi M. (1999) Polarizing microscopy of eyespot of Chlamydomonas: in situ observation of its location, orientation, and multiplication. Biospectroscopy 5, 93–100 [DOI] [PubMed] [Google Scholar]
- 19).Liu D. J., Day L. A. (1994) Pf1 virus structure: helical coat protein and DNA with paraxial phosphates. Science 265, 671–674 [DOI] [PubMed] [Google Scholar]
- 20).Tsuboi M., Kubo Y., Ikeda T., Overman S. A., Osman O., Thomas G. J., Jr (2003) Protein and DNA residue orientations in the filamentous virus Pf1 determined by polarized Raman and polarized FTIR spectroscopy. Biochemistry 42, 940–950 [DOI] [PubMed] [Google Scholar]
- 21).Tsuboi M., Overman S. A., Benevides J. M., Thomas G. J., Jr (2008) Polarized Raman Microscopy of DNA in Bacteriophage Pf1. Unpublished Results. [Google Scholar]
- 22).Allemand J. F., Bensimon D., Lavery R., Croquette V. (1998) Stretched and overwound DNA forms a Pauling-like structure with exposed bases. Proc. Natl. Acad. Sci. USA 95, 14152–14157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23).Smith G. P. (1985) Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science 228, 1315–1317 [DOI] [PubMed] [Google Scholar]
- 24).Marvin D. A., Hale R. D., Nave C., Helmer-Citterich M. (1994) Molecular models and structural comparisons of native and mutant class I filamentous bacteriophages Ff (fd, f1, M13), If1 and IKe. J. Mol. Biol. 235, 260–286 [DOI] [PubMed] [Google Scholar]
- 25).Overman S. A., Tsuboi M., Thomas G. J., Jr (1996) Subunit orientation in the filamentous virus Ff (fd, f1, M13). J. Mol. Biol. 259, 331–336 [DOI] [PubMed] [Google Scholar]
- 26).Tsuboi M., Overman S. A., Nakamura K., Rodriguez-Casado A., Thomas G. J., Jr (2003) Orientation and interactions of an essential tryptophan (Trp-38) in the capsid subunit of Pf3 filamentous virus. Biophys. J. 84, 1969–1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27).Tsuboi M., Benevides J. M., Bondre P., Thomas G. J., Jr (2005) Structural details of the thermophilic filamentous bacteriophage PH75 determined by polarized Raman microspectroscopy. Biochemistry 44, 4861–4869 [DOI] [PubMed] [Google Scholar]
- 28).Pederson D. M., Welsh L. C., Marvin D. A., Sampson M., Perham R. N., Yu M., et al. (2001) The protein capsid of filamentous bacteriophage PH75 from Thermus thermophilus. J. Mol. Biol. 309, 401–421 [DOI] [PubMed] [Google Scholar]
- 29).Blanch E. W., Hecht L., Day L. A., Pederson D. M., Barron L. D. (2001) Tryptophan absolute stereochemistry in viral coat proteins from Raman optical activity. J. Am. Chem. Soc. 123, 4863–4864 [DOI] [PubMed] [Google Scholar]
- 30).Overman S. A., Bondre P., Maiti N. C., Thomas G. J., Jr (2005) Structural characterization of the filamentous bacteriophage PH75 from Thermus thermophilus by Raman and UV-resonance Raman spectroscopy. Biochemistry 44, 3091–3100 [DOI] [PubMed] [Google Scholar]
- 31).Williams K. A., Glibowicka M., Li Z., Li H., Khan A. R., Chen Y. M., et al. (1995) Packing of coat protein amphipathic and transmembrane helices in filamentous bacteriophage M13: role of small residues in protein oligomerization. J. Mol. Biol. 252, 6–14 [DOI] [PubMed] [Google Scholar]
- 32).Marvin D. A. (1998) Filamentous phage structure, infection and assembly. Curr. Opin. Struc. Biol. 8, 150–158 [DOI] [PubMed] [Google Scholar]
- 33).Tsuboi M., Overman S. A., Thomas G. J., Jr (1996) Orientation of tryptophan-26 in coat protein subunits of the filamentous virus Ff by polarized Raman microspectroscopy. Biochemistry 35, 10403–10410 [DOI] [PubMed] [Google Scholar]
- 34).Marvin D. A., Welsh L. C., Symmons M. F., Scott W. R., Straus S. K. (2006) Molecular structure of fd (f1, M13) filamentous bacteriophage refined with respect to X-ray fibre diffraction and solid-state NMR data supports specific models of phage assembly at the bacterial membrane. J. Mol. Biol. 355, 294–309 [DOI] [PubMed] [Google Scholar]
- 35).Arnold G. E., Day L. A., Dunker A. K. (1992) Tryptophan contributions to the unusual circular dichroism of fd bacteriophage. Biochemistry 31, 7948–7956 [DOI] [PubMed] [Google Scholar]
- 36).Wen Z. Q., Thomas G. J., Jr (2000) Ultraviolet-resonance Raman spectroscopy of the filamentous virus Pf3: interactions of Trp 38 specific to the assembled virion subunit. Biochemistry 39, 146–152 [DOI] [PubMed] [Google Scholar]
- 37).Day L. A., Marzec C. J., Reisberg S. A., Casadevall A. (1988) DNA packing in filamentous bacteriophages. Annu. Rev. Biophys. Bio-phys. Chem. 17, 509–539 [DOI] [PubMed] [Google Scholar]
- 38).Tsuboi M., Ushizawa K., Nakamura K., Benevides J. M., Overman S. A., Thomas G. J., Jr (2001) Orientations of Tyr 21 and Tyr 24 in the capsid of filamentous virus Ff determined by polarized Raman spectroscopy. Biochemistry 40, 1238–1247 [DOI] [PubMed] [Google Scholar]
- 39).Fraser R. D., MacRea T. P. (1973) In “Conformation in Fibrous Proteins and Related Synthetic Polypeptides”. Academic Press, N.Y [Google Scholar]
- 40).Pauling L., Corey R. B. (1953) Two rippled-sheet configurations of polypeptide chains, and a note about the pleated sheets. Proc. Natl. Acad. Sci. USA 39, 253–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41).Yokote Y., Kubo Y., Takahashi R., Ikeda T., Akahane K., Tsuboi M. (2007) Structural details of a fowl feather elucidated by using polarized Raman microspectroscopy. B. Chem. Soc. JPN 80, 1148–1156 [Google Scholar]
- 42).Takahashi Y., Arai K. M., Yokote Y., Akahane K. (1993) A model of the polypeptide chain in the fowl feather rachis. Sci. Bull. Josai Univ. 1, 31 [Google Scholar]
- 43).Rousseau M. E., Lefevre T., Beaulieu L., Asakura T., Pezolet M. (2004) Study of protein conformation and orientation in silkworm and spider silk fibers using Raman microspectroscopy. Biomacromolecules 5, 2247–2257 [DOI] [PubMed] [Google Scholar]
- 44).Benevides J. M., Tsuboi M., Bamford J. K., Thomas G. J., Jr (1997) Polarized Raman spectroscopy of double-stranded RNA from bacteriophage phi6: local Raman tensors of base and backbone vibrations. Biophys. J. 72, 2748–2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45).Tsuboi M., Ueda T., Ushizawa K., Nagashima N. (1995) Raman tensor of adenine residue for the 1580 cm−1 vibration and its orientation in a biopolymer. J. Raman Spectrosc. 26, 745–749 [Google Scholar]
- 46).Ushizawa K., Ueda T., Tsuboi M. (1998) Polarized Raman spectrum of single crystal uridylyl (3′-5′) adenosine: Local Raman tensors of some functional groups. Biopolymers 45, 135–147 [Google Scholar]
- 47).Tsuboi M., Ueda T., Ushizawa K. (1995) Localized Raman tensors in some biopolymers. J. Mol. Struct. 352–353, 509–517 [Google Scholar]
- 48).Ueda T., Ushizawa K., Tsuboi M. (1996) Local Raman tensors of cytidine and their orientations in poly(rI)·poly(rC). J. Mol. Struct. 379, 171–187 [Google Scholar]
- 49).Kumakura A., Tsuboi M., Ushizawa K., Ueda T. (1996) Polarized Raman spectrum of a single crystal of AZT. Biospectroscopy 2, 233–242 [Google Scholar]
- 50).Ueda T., Shinozaki K., Ushizawa K., Yimit A., Tsuboi M. (1997) Raman scattering tensors of 5-iodouracil. Nucleic Acids Symp. Ser. 37, 95–96 [PubMed] [Google Scholar]
- 51).Ueda T., Shinozaki K., Ushizawa K., Tsuboi M. (1997) Raman scattering tensors of pseudouridine. J. Raman Spectrosc. 28, 947–952 [Google Scholar]
- 52).Tsuboi M., Ushizawa K., Ueda T. (1992) Assignments of Raman tensors to Raman bands of nucleic acids: use of a single crystal of adenosine triphosphoric acid and DNA fibers. Nucleic Acids Symp. Ser. 27, 61–62 [PubMed] [Google Scholar]
- 53).Ueda T., Ushizawa K., Tsuboi M. (1994) Local Raman tensors in adenosine triphosphoric acid. Spectrochim. Acta A 50, 1661–1674 [Google Scholar]
- 54).Ushizawa K., Yimit A., Ueda T., Tsuboi M. (1997) Raman scattering tensors of adenine. Nucleic Acids Symp. Ser. 37, 37–38 [PubMed] [Google Scholar]
- 55).Ushizawa K., Ueda T., Tsuboi M. (1996) Polarized micro Raman spectroscopic studies of nucleic acid: local Raman tensors in inosine-5′-monophosphoric acid and the orientation of ekypoxanthine residue in poly(rI)·poly(rC) fiber. Nucleos. Nucleot. Nucl. 15, 569–584 [Google Scholar]
- 56).Tsuboi M., Kaneuchi F., Ikeda T., Akahane K. (1991) Infrared and Raman microscopy of fowl feather barb. Can. J. Chem. 69, 1752–1757 [Google Scholar]
- 57).Tsuboi M., Kubo Y., Akahane K., Benevides J. M., Thomas G. J. (2006) Determination of the amide I Raman tensor for the antiparallel beta-sheet: application to silkworm and spider silks. J. Raman Spectrosc. 37, 240–247 [Google Scholar]
- 58).Tsuboi M., Suzuki M., Overman S. A., Thomas G. J., Jr (2000) Intensity of the polarized Raman band at 1340–1345 cm−1 as an indicator of protein alpha-helix orientation: application to Pf1 filamentous virus. Biochemistry 39, 2677–2684 [DOI] [PubMed] [Google Scholar]