Abstract
We have tested whether a gene encoding a polygalacturonase-inhibiting protein (PGIP) protects tobacco against a fungal pathogen (Rhizoctonia solani) and two oomycetes (Phytophthora parasitica var. nicotianae and Peronospora hyoscyami f. sp. tabacina). The trials were performed in greenhouse conditions for R. solani and P. parasitica and in the field for P. hyoscyami. Our results show that expression of PGIP is a powerful way of engineering a broad-spectrum disease resistance.
Keywords: disease resistance, polygalacturonase inhibitor, oomycetes, plant protection
INTRODUCTION
Plants that in nature are exposed to biotic stresses often resist to pathogen infection by rapidly activating the innate immune system. An efficient activation of the resistance responses relies on the prompt perception/transduction of signal molecules that are common to many classes of pathogens (pathogen-associated molecular patterns, PAMPs) and are recognized by germ line-encoded pattern recognition receptors (PRRs). Resistant responses are also triggered by race-specific molecules (effectors, Avr products) recognized by the so-called Resistance (R) proteins that are present in specific cultivars of several but not all crop plants (Jones and Dangl, 2006). R proteins have been widely used in breeding programs as well as for genetic transformation to protect plants against specific pathogen genotypes. Usually R-mediated resistance does not last for a long time since pathogens continually evolve more aggressive genotypes. More recently it has been proposed that a PRR-mediated recognition of PAMPs may be utilized to confer to transgenic plants a larger spectrum of disease resistance. Indeed, the Arabidopsis EFR that recognizes the bacterial elongation factor EF-Tu has been shown to confer resistance against several bacteria when transferred into Solanaceae plants (Lacombe et al., 2010). It has also been proven that chimeric PRRs may be used to engineer resistance against both bacteria or fungi (Brutus et al., 2010; De Lorenzo et al., 2011). The combination in a single plant of different PRRs as well as of chimeric PRRs that recognize several non-self structures likely represents the best way of constructing broad-spectrum and long lasting disease resistances.
The plant cell walls constitute the first line of defense against microbes. The majority of pathogenic microorganisms produce cell wall degrading enzymes (CWDEs) that are especially important for those pathogens that lack specialized penetration structures. Among the different CWDEs produced by fungi polygalacturonases (PGs) play a critical role since their action on pectin makes other cell wall components more accessible to other CWDEs (Lionetti et al., 2010). Consequently, as a strategy to optimize the action of CWDEs, PGs are often the first enzymes secreted by pathogens growing on the plant cell walls (De Lorenzo et al., 2001). PG-inhibiting proteins (PGIPs) are well-characterized proteins that recognize microbial and insect PGs and interfere with the plant cell wall degradation during pathogen attacks. These proteins are leucine-rich repeat (LRR) proteins like most of the R proteins and several PAMP receptors (Casasoli et al., 2009). They not only inhibit PGs and retard the hydrolysis of pectin but also favor the accumulation of oligogalacturonides (OGs), a class of damage-associated molecular patterns (DAMPs) that, like PAMPs, activate the plant innate immunity system (Brutus et al., 2010). The importance of PGIPs in resistance against the necrotrophic fungus Botrytis cinerea is well established: transgenic tomato and grapevine plants expressing a pear PGIP or transgenic tobacco and Arabidopsis plants expressing, respectively, bean or Arabidopsis PGIPs are more resistant to Botrytis infection in greenhouse experiments (Powell et al., 2000; Ferrari et al., 2003; Agüero et al., 2005; Manfredini et al., 2005).
Conversely, Arabidopsis plants expressing an antisense pgip gene are more susceptible to this fungus (Ferrari et al., 2006). In spite of the lower quantity of pectin in their cell wall, also monocots are protected by transgenic expression of a bean PGIP in greenhouse trials against fungi (Fusarium graminearum and Bipolaris sorokiniana; Janni et al., 2008; Ferrari et al., 2011). A negative case is represented by tomato transgenic plants expressing PvPGIP1. Due to the limited ability of PGIP1 to inhibit PGs from F. oxysporum f. sp. lycopersici, B. cinerea, and Alternaria solani the transgenic plants did not exhibit enhanced resistance against these fungi (Desiderio et al., 1997). Here, we have tested whether PvPGIP2 from Phaseolus vulgaris L. protects tobacco against an important fungal pathogen (Rhizoctonia solani) and two dangerous oomycetes (Phytophthora parasitica var. nicotianae and Peronospora hyoscyami f. sp. tabacina). Greenhouse conditions were tested for R. solani and P. parasitica var. nicotianae while field trials were carried out for P. hyoscyami f. sp. tabacina. We propose that the use of PGIP is a powerful way of engineering a broad-spectrum disease resistance.
MATERIALS AND METHODS
TRANSGENIC PLANTS
Two independent transgenic lines of Nicotiana tabacum cv. Petit Havana SR1 plants overexpressing PvPGIP2 of P. vulgaris (accession number P58822) have been used in this work and belong to the collection of plants previously characterized (Manfredini et al., 2005). The line indicated here as 2005 corresponds to the line 12.5 described in Manfredini et al. (2005). The normal phenotype of line 2.1 and its ability to revert the dwarf phenotype of transgenic tobacco plants expressing the Aspergillus niger PG II have been previously described (Capodicasa et al., 2004; Manfredini et al., 2005).
DISEASE RESISTANCE ASSAY
Inoculation with R. solani
Transgenic and control tobacco plants (N. tabacum cv. Petit Havana SR1 provided by the Tobacco Research Institute, Cuba) were grown in 6-inch pots containing black turf and rice husk (4:1) and maintained in growth chambers at 23°C. An aggressive isolate belonging to anastomosis group n. 3 of R. solani (kindly provided by the Cuban Research Institute of Plant Health and characterized by sequencing the ITS region that matched the sequence of isolate AG3, GenBank accession number HQ241274.1) was used for inoculations. The isolate was grown on potato dextrose agar at room temperature (22–25°C) for 5 days. Colonized agar plugs were removed and transferred to 250-ml Erlenmeyer flasks containing autoclaved rice grains. The pathogen was allowed to colonize the rice grains for approximately 2 weeks at room temperature and the grains were used to inoculate tobacco. Two-week-old tobacco seedlings were inoculated with approximately six grains onto the surface of the soil according to Elliott et al. (2008). Mock-inoculated untransformed plants were used as controls. Typical symptoms caused by R. solani were monitored visually at 0, 1, 2, and 3 weeks post-inoculation (wpi). Growth of R. solani on tobacco was estimated by quantitative real-time reverse transcription PCR. The extent of colonization was determined by the ratio of transcripts of the constitutively expressed actin gene (measuring the fungal biomass) to the constitutively expressed tobacco 26S rRNA gene (measuring the plant biomass) shown on a linear scale. Amplification products were sequenced and confirmed to correspond to the R. solani actin and the tobacco rRNA transcripts. Disease incidence, which included the percentage of plants exhibiting seedling death and stem rot, was evaluated after 3 wpi according to Elliott et al. (2008). The PCR product generated was sequenced and confirmed the origin. For each time point, three root samples were taken from five plants and the experiment was repeated twice. An arcsine transformation was performed on all percent incidence data before statistical analysis in order to improve homogeneity of variance. Data were analyzed by analysis of variance or general linear model procedures of SAS (SAS Institute, Cary, NC, USA). Significant difference among mean values was determined by Fisher’s least significant difference mean separation at P = 0.05.
Inoculation with P. parasitica var. nicotianae
The pathogen P. parasitica var. nicotianae race 0 used in this study belongs to the Plant Health Institute in Havana and was isolated from naturally infected tobacco plants in Havana fields. The isolate was identified and classified through sequencing the ITS region that matched the sequence of the GenBank isolate with accession number DQ059571.1. The inoculum was prepared by sterilizing and infesting toothpicks with the test organism. Toothpicks were autoclaved in V8 juice for 15 min at 121°C, allowed to cool, placed on Petri plates filled with potato dextrose agar, and inoculated with a 5-mm plug from actively growing cultures of P. parasitica var. nicotianae. Plates were incubated at 27°C in the dark for 10–14 days, allowing the fungus to grow across the plates and into the media-impregnated toothpicks.
During the greenhouse evaluation the tobacco transgenic lines and wild type plants were inoculated by aseptically pushing the infested toothpicks into stems 2–3 cm above the soil line or into root systems near the base of the plant (Sullivan et al., 2005). Greenhouse temperature was ranging from 15 to 25°C during the tests. Uninfected toothpicks acted as controls. Each treatment had 50 plants and was replicated five times. Each transgenic line and wild type plant was kept in separate trays and placed in separate float baths to prevent cross contamination during the experiment. Development of stem lesions was evaluated using a linear scale of 1–10, where 1 was no disease and 10 was a dead plant according to Csinos (1999). The ratings were taken on stems at 10 days post-inoculation. An arcsine transformation was performed on all percent incidence data before statistical analysis in order to improve homogeneity of variance. Data were subjected to analysis of variance or general linear model procedures of SAS (SAS Institute, Cary, NC, USA). Significant difference among means was determined by Fisher’s least significant difference mean separation at P = 0.05.
Resistance to P. hyoscyami f. sp. tabacina
The P. hyoscyami f. sp. tabacina isolate belongs to the Plant Health Institute in Havana and was collected from a tobacco field near Havana, identified and classified through sequencing the ITS region that matched the sequence of the GenBank isolate with accession number DQ067898.1. To determine the performance of tobacco plants expressing PvPGIP2 under field conditions, trials were conducted in the tobacco area in Havana with a high inoculum pressure where P. hyoscyami f. sp. tabacina is a significant problem for tobacco production each year. During cold and wet season of 2009, the two transgenic Pvpgip2 lines and wild type plants were evaluated by planting fifty 8-week-old plants of each line in the tobacco production area. The plants were planted with a random design to look at positional effects in the field and five replicates were made. The percentages of healthy plants were determined, where more than three blue molds spot per leaf was considered as an unhealthy plant at 35 days post-planting. An arcsine transformation was performed on all percent incidence data before statistical analysis in order to improve homogeneity of variance. Data were subjected to analysis of variance or general linear model procedures of SAS (SAS Institute, Cary, NC, USA). Significant difference among means was determined by Fisher’s least significant difference mean separation at P = 0.05.
RESULTS
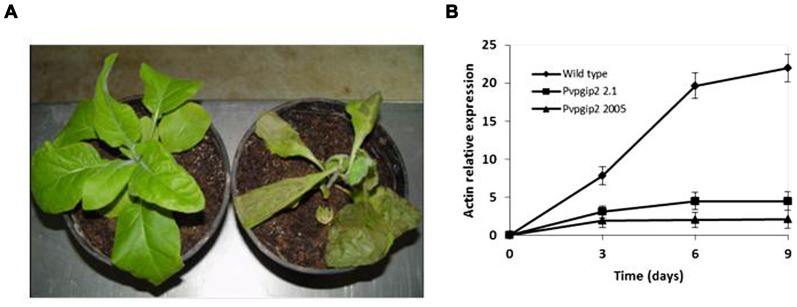
One of the genes of common bean (Pvpgip2) encodes the most efficient and wide-spectrum PG inhibitor so far studied (Casasoli et al., 2009). To assess the effectiveness of PvPGIP2 in protecting tobacco against R. solani, P. parasitica var. nicotianae, and P. hyoscyami f. sp. tabacina, two transgenic lines were evaluated. Under greenhouse conditions, the main symptoms caused by R. solani on wild type tobacco were small stem water-soaked lesions that rapidly become brown and sunken, primarily at the level of the soil line or closely above it. Lesions subsequently expanded throughout the stems causing the tissue to turn brown and die. Disease symptoms (both seedling death and stem rot) were severe on wild type plants and very limited and less visible in transgenic lines (Table 1). The two transgenic lines behaved similarly (line 2.1 is shown in Figure 1A as a representative result). Symptom development coincided with an increase of fungal biomass in the colonized roots of the wild type plants while no significant increase of fungal biomass occurred in transgenic plants (Figure 1B).
Table 1.
Reaction of tobacco plants expressing Pvpgip2 to R. solani in greenhouse conditions.
| Genotype name | Disease incidence (%)a | |
|---|---|---|
| Seedling deathb | Stem rot | |
| Nicotiana tabacum cv. SR1 expressing Pvpgip2 line 2.1 | 16.3* | 14.2* |
| Nicotiana tabacum cv. SR1 expressing Pvpgip2 line 2005 | 15.8* | 17.2* |
| Nicotiana tabacum cv. SR1 | 42.4§ | 52.8§ |
| CV (%)c | 6.2 | 7.1 |
Arcsine-transformed percentage of disease incidence.
Seedling death was measured at 3 weeks post-inoculation.
Coefficient of variation (N = 50). Values designated with the same symbol are not significantly different (P > 0.05).
FIGURE 1.
Greenhouse evaluation of tobacco transgenic lines expressing the Pvpgip2 gene inoculated with R. solani. Inoculated transgenic line Pvpgip2 2.1 (A) and wild type tobacco plants at 3 wpi. (B) Quantitative RT-PCR measuring R. solani growth in tobacco transgenic lines and wild type plants. Accumulation of fungal actin PCR transcripts was determined using tobacco 26S rRNA gene as a reference. Each point represents mean values with standard error (N = 5). The photographs were obtained at 50 cm of distance.
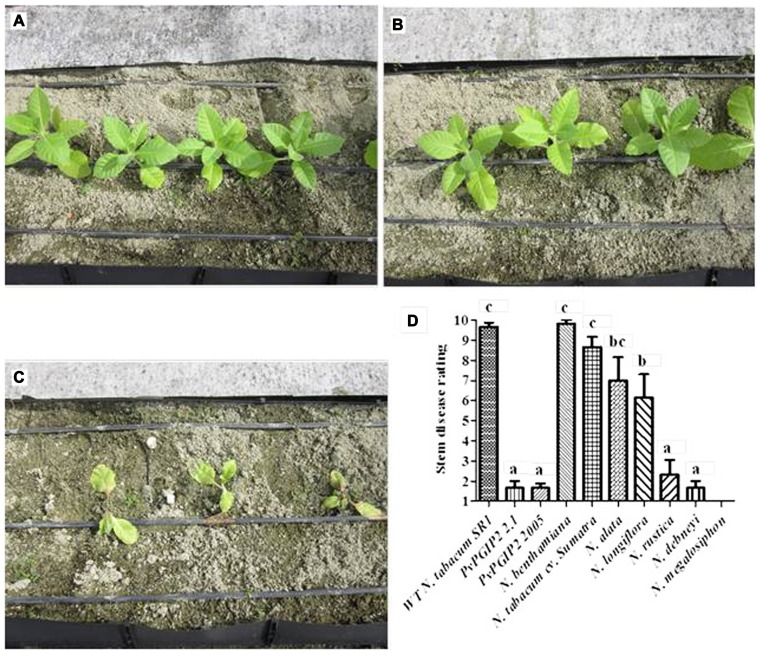
Under greenhouse conditions the two transgenic tobacco lines expressing Pvpgip2 were also remarkably resistant to the oomycete pathogen P. parasitica var. nicotianae. At 2-week post-inoculation slight disease symptoms appeared on the wild type plants whereas no symptoms were detected on the transgenic plants. However, at 5-week post-inoculation severe disease symptoms (leaf wilting and stem rot) were observed in the wild type plants. All wild type plants died 5 days later. Instead both transgenic lines expressing Pvpgip2 remained healthy and showed a level of resistance similar to Nicotiana species that are naturally highly resistant to P. parasitica var. nicotianae (Figures 2A–D).
FIGURE 2.
Evaluation of Pvpgip2 transgenic tobacco plants for resistance against P. parasitica var. nicotianae. Phenotype of transgenic tobacco lines Pvpgip2 2.1 (A), Pvpgip2 2005 (B), and wild type plants (C) on a highly infected soil at 10 days after planting. The development of stem lesions was evaluated using a linear scale of 1–10, where 1 was no disease and 10 was a dead plant according to Csinos (1999). Comparative evaluation of resistance in two transgenic homozygous lines expressing PvPGIP2 and reference genotypes of Nicotiana plants with different degree of resistance in greenhouse conditions (D). Bars represent mean values with standard error (N = 50). Columns designated with the same letter are not significantly different (P > 0.05). The photographs were obtained at 100 cm of distance and are representative of one typical event. Nicotiana rustica (PI 499174), Nicotiana alata (PI 42334), Nicotiana longiflora (PI 555533), Nicotiana benthamiana (PI 555478), Nicotiana debneyi (PI 503320), Nicotiana megalosiphon (PI 555536).
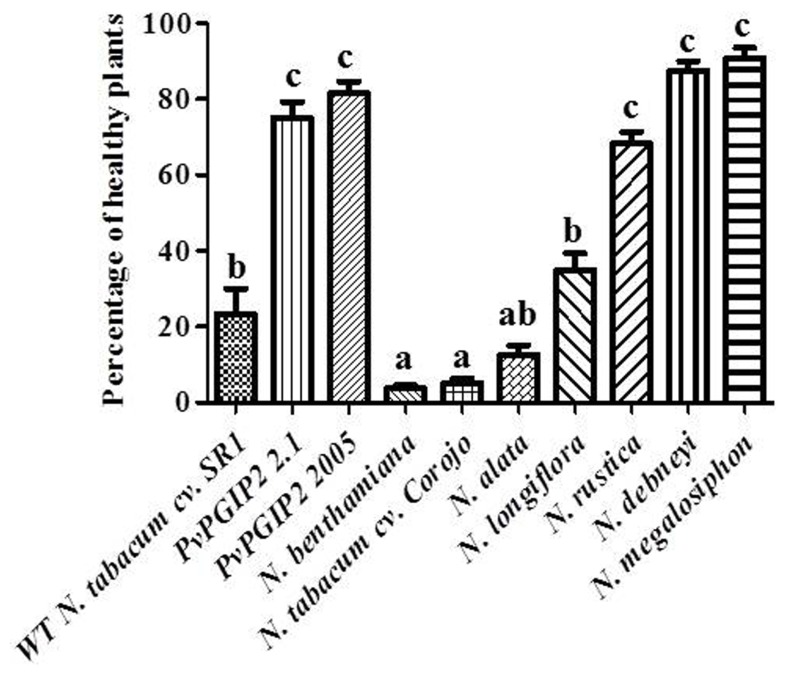
Trials were also conducted in the field during the cold and wet season when tobacco blue mold caused by P. hyoscyami f. sp. tabacina constitutes a significant problem in Cuba. Transgenic plants displayed a high level of resistance that was comparable to that of Nicotiana species that are naturally highly resistant to P. hyoscyami f. sp. tabacina (Figure 3).
FIGURE 3.
Field trials of Pvpgip2 transgenic tobacco plants for resistance against P. hyoscyami f. sp. tabacina. Quantitative evaluation of resistance to P. hyoscyami f. sp. tabacina in different transgenic lines and reference genotypes. Nicotiana plants exhibiting different degree of natural resistance at 35 days after planting. Bars represent mean values with standard error (N = 50). Columns designated with the same letter are not significantly different (P > 0.05).
DISCUSSION
We have shown that the expression of a PGIP gene from common bean in tobacco, i.e., a plant belonging to the economically important family of Solanaceae, confers to transgenic plants a strong resistance against fungi and oomycetes, both in greenhouses and in the field (P. hyoscyami f. sp. tabacina). Oomycetes are a group of eukaryotic microorganisms that includes some of the most important pathogens of plants. Among these, members of the genus Phytophthora cause enormous economic losses on crop species as well as environmental damages in natural ecosystems (Kamoun, 2006). PGs are produced by Phytophthora spp. and R. solani and are encoded by gene families (Marcus et al., 1986; Götesson et al., 2002; Wu et al., 2008). The PG–PGIP interaction has been characterized in R. solani (Akhgari et al., 2012) while many PG genes have been cloned from Phytophthora spp., but their interaction with PGIP2 of P. vulgaris has not yet been analyzed. However it is possible that the PG–PGIP interaction characterized in vitro does not reflect the situation in vivo. For example, as reported by Joubert et al. (2007), transgenic tobacco plants expressing grapevine PGIP are protected against B. cinerea without any evidence of PG–PGIPinteraction in vitro. It is possible that protection is due to effects other than the inhibition of pathogen PGs, caused by the overexpression of PGIP. PGIP may bind the most exposed and vulnerable positions in the pectin and indirectly protect pectin against degradation (Spadoni et al., 2006). On the other hand, it has been reported that transgenic plants expressing PGIP exhibit altered regulation of cell wall-associated genes; thus, the consequent cell wall modifications may be responsible for the enhanced resistance (Alexandersson et al., 2011).
The expression of a PG inhibitor neither alters the physiological performances nor exhibits detrimental effects on the growth of transgenic plants (Desiderio et al., 1997; Powell et al., 2000; Capodicasa et al., 2004; Manfredini et al., 2005; Alexandersson et al., 2011; Mohammadzadeh et al., 2012). It is therefore possible to engineer disease resistance in crop plants by using PGIPs as gene tools. The structure of PGIPs and of microbial PGs is being deeply studied; it is known, for example, that the change of one or a few residues confers to the inhibitor new recognition specificities and may improve its inhibitory strength (Leckie et al., 1999; Federici et al., 2001; Di Matteo et al., 2003; Casasoli et al., 2009; Benedetti et al., 2011). This knowledge may help in planning mutational strategies aimed at improving the properties of the natural PGIPs and their recognition versatility against the many microbial PGs evolved in nature.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by ERC AdvGrant FUEL-PATH 233083 and by Istituto Pasteur-Fondazione Cenci Bolognetti.
REFERENCES
- Agüero C. B., Uratsu S. L., Greve C., Powell A. L. T., Labavitch J. M., Meredith C. P., et al. (2005). Evaluation of tolerance to Pierce’s disease and Botrytis in transgenic plants of Vitis vinifera L. expressing the pear PGIP gene. Mol. Plant Pathol. 6 43–51 [DOI] [PubMed] [Google Scholar]
- Akhgari A. B., Motallebi M., Zamani M. R. (2012). Bean polygalacturonase-inhibiting protein expressed in transgenic Brassica napus inhibits polygalacturonase from its fungal pathogen Rhizoctonia solani. Plant Prot. Sci. 48 1–9 [Google Scholar]
- Alexandersson E., Becker J. V., Jacobson D., Nguema-Ona E., Steyn C., Denby K. J., et al. (2011). Constitutive expression of a grapevine polygalacturonase-inhibiting protein affects gene expression and cell wall properties in uninfected tobacco. BMC Res. Notes 4 493 10.1186/1756-0500-4-493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti M., Leggio C., Federici L., De Lorenzo G., Pavel N. V., Cervone F. (2011). Structural resolution of the complex between a fungal polygalacturonase and a plant polygalacturonase-inhibiting protein by small-angle X-ray scattering. Plant Physiol. 157 599–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutus A., Sicilia F., Macone A., Cervone F, De Lorenzo G. (2010). A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc. Natl. Acad. Sci. U.S.A. 107 9452–9457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capodicasa C., Vairo D., Zabotina O., McCartney L., Caprari C., Mattei B., et al. (2004). Targeted modification of homogalacturonan by transgenic expression of a fungal polygalacturonase alters plant growth. Plant Physiol. 135 1294–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casasoli M., Federici L., Spinelli F., Di Matteo A., Vella N., Scaloni F., et al. (2009). Integration of evolutionary and desolvation energy analysis identifies functional sites in a plant immunity protein. Proc. Natl. Acad. Sci. U.S.A. 106 7666–7671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csinos A. (1999). Stem and root resistance to tobacco black shank. Plant Dis. 83 777–780 [DOI] [PubMed] [Google Scholar]
- De Lorenzo G., Brutus A., Savatin D. V., Sicilia F., Cervone F. (2011). Engineering plant resistance by constructing chimeric receptors that recognize damage-associated molecular patterns (DAMPs). FEBS Lett. 585 1521–1528 [DOI] [PubMed] [Google Scholar]
- De Lorenzo G., D’Ovidio R., Cervone F. (2001). The role of polygalacturonase-inhibiting proteins (PGIPs) in defense against pathogenic fungi. Annu. Rev. Phytopathol. 39 313–335 [DOI] [PubMed] [Google Scholar]
- Desiderio A., Aracri B., Leckie F., Mattei B., Salvi G., Tigelaar H., et al. (1997). Polygalacturonase-inhibiting proteins (PGIPs) with different specificities are expressed in Phaseolus vulgaris. Mol. Plant Microbe Interact. 10 852–860 [DOI] [PubMed] [Google Scholar]
- Di Matteo A., Federici L., Mattei B., Salvi G., Johnson K. A., Savino C., et al. (2003). The crystal structure of PGIP (polygalacturonase-inhibiting protein), a leucine-rich repeat protein involved in plant defense. Proc. Natl. Acad. Sci. U.S.A. 100 10124–10128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott P. E., Lewis R. S., Shew H. D., Gutierrez W. A., Nicholson J. S. (2008). Evaluation of tobacco germplasm for seedling resistance to stem rot and target spot caused by Thanatephorus cucumeris. Plant Dis. 92 425–430 [DOI] [PubMed] [Google Scholar]
- Federici L., Caprari C., Mattei B., Savino C., Di Matteo A., De Lorenzo G., et al. (2001). Structural requirements of endopolygalacturonase for the interaction with PGIP (polygalacturonase-inhibiting protein). Proc. Natl. Acad. Sci. U.S.A. 98 13425–13430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S., Galletti R., Vairo D., Cervone F, De Lorenzo G. (2006). Antisense expression of the Arabidopsis thaliana AtPGIP1 gene reduces polygalacturonase-inhibiting protein accumulation and enhances susceptibility to Botrytis cinerea. Mol. Plant Microbe Interact. 19 931–936 [DOI] [PubMed] [Google Scholar]
- Ferrari S., Sella L., Janni M., De Lorenzo G., Favaron F, D’Ovidio R. (2011). Transgenic expression of polygalacturonase-inhibiting proteins in Arabidopsis and wheat increases resistance to the flower pathogen Fusarium graminearum. Plant Biol. 14 31–38 [DOI] [PubMed] [Google Scholar]
- Ferrari S., Vairo D., Cervone F., Ausubel F. M, De Lorenzo G. (2003). Tandemly duplicated Arabidopsis genes that encode polygalacturonase-inhibiting proteins are regulated coordinately by different signal transduction pathways in response to fungal infection. Plant Cell 15 93–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götesson A., Marshall J. S., Jones D. A., Hardham A. R. (2002). Characterization and evolutionary analysis of a large polygalacturonase gene family in the oomycete plant pathogen Phytophthora cinnamomi. Mol. Plant Microbe Interact. 15 907–921 [DOI] [PubMed] [Google Scholar]
- Janni M., Sella L., Favaron F., Blechl A. E., De Lorenzo G, D’Ovidio R. (2008). The expression of a bean PGIP in transgenic wheat confers increased resistance to the fungal pathogen Bipolaris sorokiniana. Mol. Plant Microbe Interact. 21 171–177 [DOI] [PubMed] [Google Scholar]
- Jones J. D., Dangl J. L. (2006). The plant immune system. Nature 444 323–329 [DOI] [PubMed] [Google Scholar]
- Joubert D. A., Kars I., Wagemakers L., Bergmann C., Kemp G., Vivier M. A., et al. (2007). A polygalacturonase-inhibiting protein from grapevine reduces the symptoms of the endopolygalacturonase BcPG2 from Botrytis cinerea in Nicotiana benthamiana leaves without any evidence for in vitro interaction. Mol. Plant Microbe Interact. 4 392–402 [DOI] [PubMed] [Google Scholar]
- Kamoun S. (2006). A catalogue of the effector secretome of plant pathogenic oomycetes. Annu. Rev. Phytopathol. 44 41–60 [DOI] [PubMed] [Google Scholar]
- Lacombe S., Rougon-Cardoso A., Sherwood E., Peeters N., Dahlbeck D., van Esse H. P., et al. (2010). Interfamily transfer of a plant pattern-recognition receptor confers broad-spectrum bacterial resistance. Nat. Biotechnol. 28 365–369 [DOI] [PubMed] [Google Scholar]
- Leckie F., Mattei B., Capodicasa C., Hemmings A., Nuss L., Aracri B., et al. (1999). The specificity of polygalacturonase-inhibiting protein (PGIP): a single amino acid substitution in the solvent-exposed b-strand/b-turn region of the leucine-rich repeats (LRRs) confers a new recognition capability. EMBO J. 18 2352–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionetti V., Francocci F., Ferrari S., Volpi C., Bellincampi D., Galletti R., et al. (2010). Engineering the cell wall by reducing de-methyl-esterified homogalacturonan improves saccharification of plant tissues for bioconversion. Proc. Natl. Acad. Sci. U.S.A. 107 616–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfredini C., Sicilia F., Ferrari S., Pontiggia D., Salvi G., Caprari C., et al. (2005). Polygalacturonase-inhibiting protein 2 of Phaseolus vulgaris inhibits BcPG1, a polygalacturonase of Botrytis cinerea important for pathogenicity, and protects transgenic plants from infection. Physiol. Mol. Plant Pathol. 67 108–115 [Google Scholar]
- Marcus L., Barash I., Sneh B., Koltin Y., Finkler A. (1986). Purification and characterization of pectolytic enzymes produced by virulent and hypovirulent isolates of Rhizoctonia solani Kuhn. Physiol. Mol. Plant Pathol. 29 325–336 [Google Scholar]
- Mohammadzadeh R., Zamani M., Motallebi M., Norouzi P., Jourabchi E., Benedetti M., et al. (2012). Agrobacterium tumefaciens-mediated introduction of polygalacturonase inhibiting protein 2 gene (PvPGIP2) from Phaseolus vulgaris into sugar beet (Beta vulgaris L.). Aust. J. Crop Sci. 6 1290–1297 [Google Scholar]
- Powell A. L., van Kan J., ten Have A., Visser J., Greve L. C., Bennett A. B., et al. (2000). Transgenic expression of pear PGIP in tomato limits fungal colonization. Mol. Plant Microbe Interact. 13 942–950 [DOI] [PubMed] [Google Scholar]
- Spadoni S., Zabotina O., Di Matteo A., Mikkelsen J. D., Cervone F., De Lorenzo G., et al. (2006). Polygalacturonase-inhibiting protein interacts with pectin through a binding site formed by four clustered residues of arginine and lysine. Plant Physiol. 141 557–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M. J., Melton T. A., Shew H. D. (2005). Fitness of races 0 and 1 of Phytophthora parasitica var. nicotianae. Plant Dis. 89 1220–1228 [DOI] [PubMed] [Google Scholar]
- Wu C. H., Yan H. Z., Liu F. L., Liou R. F. (2008). Functional characterization of a gene family encoding polygalacturonases in Phytophthora parasitica. Mol. Plant Microbe Interact. 21 480–489 [DOI] [PubMed] [Google Scholar]