Abstract
Influenza A virus vaccines currently contain a mixture of isolates that reflect the genetic and antigenic characteristics of the currently circulating strains. This study was conducted to evaluate the efficacy of a trivalent inactivated swine influenza virus vaccine (Flusure XP) in pigs challenged with a contemporary α-cluster H1N1 field isolate of Canadian swine origin. Pigs were allocated to vaccinated, placebo, and negative-control groups and monitored for respiratory disease for 5 d after challenge. On the challenge day and 5 d after challenge the serum of the vaccinated pigs had reciprocal hemagglutination inhibition antibody titers 40 for all the vaccine viruses but ≤ 20 for the challenge virus. Gross lesions were present in the lungs of all pigs that had been inoculated with the challenge virus, but the proportion of lung tissue consolidated did not differ significantly between the placebo and vaccinated pigs. However, the amount of virus was significantly reduced in the nasal secretions, lungs, and bronchoalveolar lavage fluid in the vaccinated pigs compared with the placebo pigs. These results indicate that swine vaccinated with Flusure XP were partially protected against experimental challenge with a swine α-cluster H1N1 virus that is genetically similar to viruses currently circulating in Canadian swine.
Résumé
Les vaccins actuels contre l’influenza A contiennent un mélange d’isolats qui reflète les caractéristiques génétiques et antigéniques des souches actuellement en circulation. La présente étude a été réalisée afin d’évaluer l’efficacité d’un vaccin inactivé trivalent contre le virus de l’influenza porcin (Flusure XP) chez des porcs challengés avec un isolat terrain du virus de l’influenza de la grappe α du type H1N1 provenant d’un porc d’origine canadienne. Des porcs ont été répartis dans un des trois groupes suivants : vacciné, placebo ou témoin négatif; et examinés pour problèmes respiratoires pendant 5 jours après le challenge. Le jour du challenge et le 5e jour suivant le challenge, on retrouvait dans le sérum des porcs vaccinés des titres réciproques d’anticorps hémagglutinants 40 pour tous les virus vaccinaux mais ≤ 20 pour le virus ayant servi au challenge. Des lésions macroscopiques étaient présentes dans les poumons de tous les porcs qui avaient été inoculés avec le virus servant pour le challenge, mais il n’y avait pas de différence significative dans la proportion de tissu pulmonaire consolidé entre le groupe vacciné et le groupe placebo. Toutefois, la quantité de virus était réduite de manière significative dans les sécrétions nasales, les poumons et le liquide des lavages broncho-alvéolaires des porcs vaccinés comparativement aux porcs du groupe placebo. Ces résultats indiquent que les porcs vaccinés avec Flusure XP étaient partiellement protégés contre une infection expérimentale avec un virus H1N1 porcin de la grappe α qui est génétiquement similaire aux virus qui circulent actuellement chez les porcs canadiens.
(Traduit par Docteur Serge Messier)
Introduction
Influenza A viruses (IAVs) are a major cause of acute respiratory disease in swine that occurs year-round and has a significant economic impact on production (1). The predominant IAV subtypes found in North American swine are H1N1, H1N2, and H3N2. Within these subtypes the viruses can be further divided into related groups by the genetic and antigenic properties of the hemagglutinin (HA) gene. Vaccines are one of the principal measures used to mitigate the effects of IAV infection in pigs. The vaccines that are commercially available for use in swine in the United States and Canada contain a mixture of 2 or more inactivated viruses with genetic and antigenic characteristics of the currently circulating strains. Since the predominant antibody response to these vaccines is to the HA proteins of the viruses (2), the cross-reactive properties of the antibodies to the HA proteins in the vaccines and the genetic relationships of IAVs in swine are used to evaluate the vaccine viruses for potential cross-reactivity and protection.
Within the North American swine HA subtype 1 viruses (SwH1), phylogenetic and antigenic analyses have revealed 4 distinct genetic clusters, referred to as α, β, γ, and δ (3,4). The α, β, and γ clusters share ancestry with classic swine influenzavirus isolates such as the prototypical A/swine/Iowa/15/1930 (H1N1) (3). The δ cluster viruses are most closely related to human seasonal IAVs and upon further investigation have been found to have 2 distinct subclusters, δ1 and δ2, which can have substantial differences in all 8 viral genes, likely the result of 2 separate introductions from humans to pigs (4). Owing to their different ancestry, there is variable antigenic cross-reactivity between the α, β, and γ clusters but limited to no cross-reactivity between the δ viruses and the other 3 clusters (3,4). The Canadian human-like swine viruses are in the δ1 subcluster, and the US human-like swine viruses are in both subclusters (4,5). The 2009 pandemic virus (pH1N1) possesses an HA gene similar to that of the SwH1γ cluster and was the predominant virus circulating in humans and pigs in North America during the pandemic and immediate postpandemic periods (MRG and SED: unpublished data). Before the introduction of pH1N1 to pigs in Canada and the United States the predominant strains circulating in US swine were SwH1β, -γ, and -δ (6), whereas SwH1α and SwH1δ predominated in Canadian swine (7). For this reason, an SwH1α virus was selected for the challenge strain.
The vaccines that are commercially available in North America contain inactivated IAVs representing the subtypes and genetic clusters currently circulating in North America. Although the cross-reactivity of hemagglutination inhibition (HI) antibodies and analysis of HA gene similarity are used as predictors of vaccine cross-protection, challenge studies in pigs using heterologous contemporary field isolates are the gold standard for evaluating vaccine efficacy against new strains. The present study was conducted to evaluate the efficacy of a vaccine containing an SwH1γ virus [A/swine/Iowa/110600/2000 (H1N1)] in pigs challenged with a contemporary SwH1α field isolate of Canadian swine origin, A/swine/Illinois/02450/2008 (H1N1).
Materials and methods
Viruses and sequence analyses
The commercially available vaccine used for this experiment, FluSure XP (Pfizer Animal Health, Madison, New Jersey, USA), contained 3 influenza viruses: A/swine/Iowa/110600/2000 (H1N1) (IA00 H1N1), A/swine/North Carolina/031/2005 (H1N1) (NC05 H1N1), and A/swine/Missouri/069/2005 (H3N2) (MO05 H3N2). This is the formulation available in Canada (the product available in the United States also contains A/swine/Oklahoma/0726H/2008). The challenge virus, influenza A/swine/Illinois/02450/2008 (H1N1) (IL08 H1N1), was selected from an IAV archive at the University of Minnesota Veterinary Diagnostic Laboratory (UMVDL), St. Paul, Minnesota, USA. The virus had originally been isolated from a lung tissue sample collected postmortem from an 11-week-old pig that had severe respiratory disease. Sequencing of the HA gene was done on the original sample submission by means of a viral RNA extraction kit (Applied Biosystems, Foster City, California, USA), a combination of universal (8) and custom-made primers (available upon request), and a genetic analyzer (Applied Biosystems). Phylogenetic analyses, molecular evolutionary analyses, and antigenic analyses were conducted with the use of MEGA 4 (9). For the sequence alignments, we used IAV sequences available in the US National Center for Biotechnology Information’s GenBank and the UMVDL database, with the aid of Clustal W and the phylogenetic analyses. The evolutionary history was inferred by means of the neighbor-joining method, and evolutionary distances were computed with the maximum composite likelihood method and the Kimura 2-parameter model (9–12). Putative antigenic sites in the HA gene were identified by alignment to A/Puerto Rico/8/1934 (13,14). The virus inoculum was prepared by propagation on Madin–Darby canine kidney (MDCK) cells (15) and had a titer of 105.5 TCID50 (median tissue culture infective dose)/mL, as calculated by the Spearman–Kärber method (16).
Serologic study
Serum samples were pretreated with receptor-destroying enzyme (RDE) and turkey erythrocytes (17). Serial 2-fold dilutions of the treated samples, from 1:10 to 1:640, were assayed for anti-influenza A antibodies by the standard HI test with 4 to 8 hemagglutination units of virus and a 0.5% suspension of turkey erythrocytes (17). The HI tests were conducted against the challenge virus (IL08 H1N1) and the vaccine viruses (IA00 H1N1, NC05 H1N1, and MO05 H3N2).
Experimental design
The animal phase of the study was conducted with the approval of the University of Minnesota Institutional Care and Use Committee. Twenty-four 3-week-old pigs were purchased from Manthei Hog Farm, Elk River, Minnesota, USA, and housed in an isolation facility at the University of Minnesota equipped with negative-pressure air flow and individual-room air filtration. All the pigs were clinically healthy upon arrival, free from infection with IAVs, Mycoplasma hyopneumoniae, and porcine reproductive and respiratory syndrome virus (PRRSV), and had no serum antibodies against IAVs. All were treated with a single intramuscular dose of ceftiofur crystalline-free acid (Excede; Pfizer Animal Health, Kalamazoo, Michigan, USA) according to label instructions before being shipped to reduce bacterial contamination. The animals were randomly assigned to treatment with a generalized block design that was based on body weight and placed in assigned isolation rooms upon arrival at the isolation facility (Table I). The pigs were given free access to water and were fed ad libitum an antibiotic-free, age-appropriate diet that met or exceeded US National Research Council nutrient requirements.
Table I.
Summary of the experimental design
| Room number and group ID | Treatmenta | Number of swine | Challengeb |
|---|---|---|---|
| Room 1, NTX | No treatment | 2 | None |
| Room 2 | |||
| NTX | No treatment | 1 | Nonec |
| T01 | Vaccination | 5 | SwH1α |
| T02 | Given adjuvant alone | 5 | SwH1α |
| Room 3 | |||
| NTX | No treatment | 1 | Nonec |
| T01 | Vaccination | 4 | SwH1α |
| T02 | Given adjuvant alone | 4 | SwH1α |
A trivalent vaccine containing A/swine/Iowa/110600/2000 (H1N1) (IA00 H1N1), A/swine/North Carolina/031/2005 (H1N1) (NC05 H1N1), and A/swine/Missouri/069/2005 (H3N2) (MO05 H3N2) was given as 2 injections of 2 mL 14 d apart.
Two weeks after the 2nd vaccination the pigs were challenged with A/swine/Illinois/02450/2008 (H1N1), a contemporary field isolate of Canadian swine origin. The inoculum had a virus titer of 105.5 TCID50 (median tissue culture infective dose)/mL; 2 mL was injected intratracheally and 2 mL intranasally.
Sentinel pigs removed for euthanasia and necropsy before challenge.
On experimental day 0 the pigs in groups T01 and T02 received 2 mL of vaccine adjuvant alone or reconstituted vaccine, respectively, intramuscularly on the left side of the neck. The intramuscular injections were repeated 14 d later (day 14) on the right side of the neck. The pigs in group NTX (n = 4) were not treated and served as the negative controls: 2 were housed in a separate room until the end of the study, and the other 2 served as sentinel pigs. Immediately before the virus challenge the sentinel pigs, which were housed in rooms 2 and 3 with the vaccinated pigs, were removed for euthanasia and necropsy to demonstrate the health status of the pigs before challenge. On experimental day 28 (2 wk after the 2nd vaccination), all the pigs in groups T01 and T02 were inoculated with IL08 H1N1, 2 mL intratracheally and 2 mL intranasally, while anesthetized with a single intramuscular dose (6.6 mg/kg) of tiletamine/zolazepam (Telazol; Fort Dodge Animal Health, Fort Dodge, Iowa, USA). Two pigs were removed from the study because of a treatment allocation error, which left 9 pigs per treatment group.
Clinical and pathological examination
The personnel conducting the clinical and injection-site observations, necropsy, and laboratory analysis of samples collected were blinded to the treatments given to the pigs. The pigs were examined daily throughout the study by a veterinarian for signs of lethargy, changes in respiration, appetite, injection-site reactions, and other changes in general health. The pigs were also examined for coughing, labored breathing, and nasal discharge from 1 day before challenge (day 27) until the end of the study (day 33). The rectal temperature was measured and recorded starting 2 d before challenge (day 26) until the end of the study. A nasal swab was collected 1 day before challenge to confirm the negative status at challenge and then every 24 h after challenge (days 29 to 33). Serum was collected 12 and 2 d before the 1st vaccination (days −12 and −2), 1 day before the 2nd vaccination (day 13), at challenge (day 28), and 5 d after challenge (day 33). On day 33 the pigs were euthanized for necropsy.
Euthanasia of the 2 sentinel pigs and the remaining 18 treated pigs was by means of a lethal dose of pentobarbital (Fatal-Plus Solution; Vortech Pharmaceuticals, Dearborn, Michigan, USA). All necropsies were conducted immediately after euthanasia. The lungs were removed with the trachea attached and evaluated for the percentage of the lung affected with purple, lobular consolidation typical of influenza infection in swine. The percentage consolidation for each lobe was calculated from weighted proportions of the total lung volume (18). The proportions were 10% each for the cranial, middle, and accessory lobes and 25% each for the caudal lobes. A bronchoalveolar lavage fluid (BALF) sample was collected for virus isolation by clamping the cranial trachea, searing the outside of the caudal trachea with a hot spatula, and using a sterile 14-gauge needle to inject 50 mL of Eagle’s minimum essential medium (MEM) containing 4% bovine serum albumin, trypsin, and antibiotics (MEM+) into the lungs. After gentle massage of the lungs, approximately 25 mL of fluid was aspirated with the same needle and syringe. Two samples of the right middle lung lobe (or lesion, if present) were collected; one was tested for the presence of virus by reverse-transcription polymerase chain reaction (RT-PCR) and by virus isolation, and the other was fixed in 10% formalin for 48 h and embedded in paraffin for histopathological study by standard techniques. The lung tissue was examined microscopically for bronchiolar epithelial changes and peribronchiolar inflammation in large, medium, and small bronchioles. The following scoring stratagem was used: 0 — no airways affected; 1 — only a few isolated airways affected; 2 — localized cluster of affected airways (lobular); 3 — several airways affected throughout the section; and 4 — many airways affected, often severely (19). The scores were categorized for statistical analysis as 0–2 and 3–4.
Virus isolation and titration
Within 24 h of collection, the nasal swabs were placed in a vial containing 2 mL of MEM+, vortexed for 30 s, and then centrifuged for 30 min at 2272 × g. Lung tissue was homogenized with MEM+ by means of a Stomacher (Seward Laboratory Systems, Bohemia, New York, USA) for 30 s and then centrifuged for 60 min at 2272 × g.
The supernatant of the swabs collected before challenge (day 27) was tested by a previously described matrix real-time RT-PCR procedure (20,21) using magnetic bead RNA extraction (Ambion MagMAX, Applied Biosystems) and a real-time RT-PCR kit (Qiagen, Valencia, California, USA).
Virus isolation was conducted on all nasal swabs collected after challenge that were positive by the matrix RT-PCR procedure, as well as all lung and BALF samples, by inoculation of the swab and lung supernatant and the BALF on monolayers of MDCK cells grown in flasks with MEM+ (15). All cultures were incubated at 37°C in a 5% CO2 atmosphere and examined daily for 7 d for cytopathic effects with an inverted light microscope. Ten-fold serial dilutions of the positive samples were inoculated into 96-well plates containing monolayers of MDCK cells, 4 wells per dilution. After 4 d of incubation the plates were assessed for cytopathic effects and virus titers calculated by the Spearman–Kärber method (16).
All swab and lung supernatant and BALF samples found to be negative by virus isolation were retested by the matrix RT-PCR procedure. Sequencing of the HA gene, with the procedure described above, was conducted on virus isolated from the nasal swabs of 1 placebo pig and 1 vaccinated pig on day 32 (4 d after challenge) by extracting virus RNA from the MDCK cell fluids.
Additional diagnostic testing was done to determine the status of each pig for North American and European (NAEU) variants of PRRSV, Porcine circovirus-2 (PCV-2), M. hyopneumoniae, and bacterial pathogens. For each pig, tissue pools of tonsil, lung, mesenteric lymph node, and spleen were prepared for PCR tests, a bronchial swab was collected for the M. hyopneumoniae PCR test, and lung tissue was collected for aerobic culture. These tests were done at the UMVDL using protocols approved by the US Department of Agriculture.
Statistical analysis
The rectal temperatures, logarithm-transformed virus titers for nasal swabs, and logarithm-transformed antibody titers were evaluated by a general linear repeated-measures mixed model with the use of SAS software (SAS Institute, Cary, North Carolina, USA). The percentage of total lung with lesions, logarithm-transformed virus titers from BALF and lung tissue, and logarithm-transformed area under the curve (AUC) of virus titers for nasal swabs were analyzed with a general linear mixed model. Microscopic lung lesion scores categorized as 0–2 or 3–4 were analyzed by means of a generalized linear mixed model with a logit link function and binomial distribution as well as an overdispersion parameter. A P-value of < 0.05 was considered to indicate a statistically significant difference between groups. All hypothesis tests were 2-tailed. Data collected from the sentinel pigs, which were euthanized before inoculation, were not used in the statistical analysis.
Results
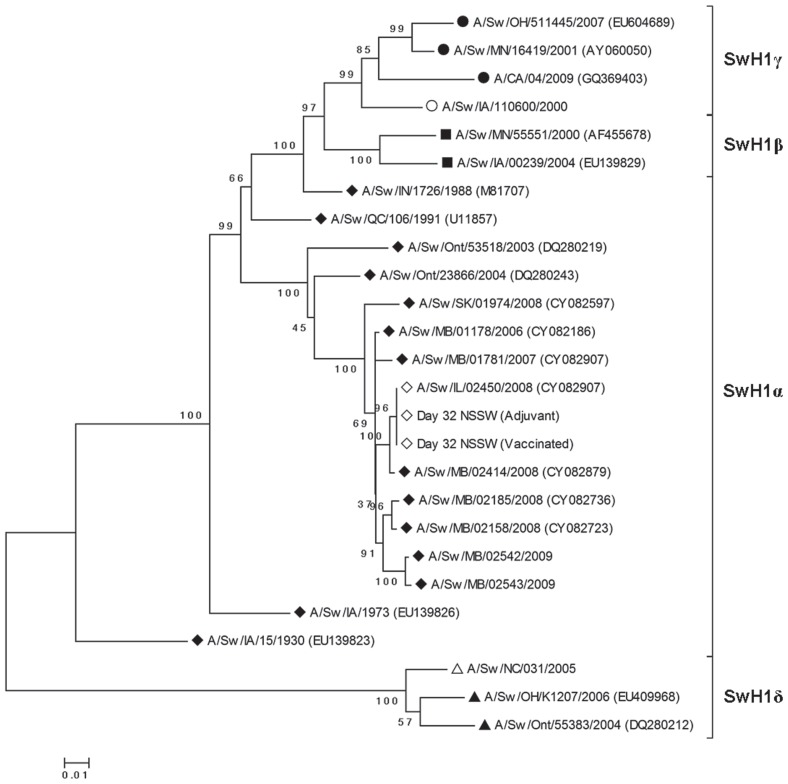
The evolutionary history of IL08 (H1N1) was inferred from the phylogenetic analysis of the HA gene sequences. The virus clustered tightly within the swine HA type 1 α cluster (SwH1α) with isolates from Manitoba (Figure 1). In addition to the genetic similarity to viruses of Canadian swine origin, there was an epidemiologic link, as the pigs in Illinois were geographically co-located with pigs imported from Canada. Pairwise distance analysis revealed that the challenge virus had 99.7% nucleic acid similarity to A/swine/Manitoba/02414/2008 and 99.1% similarity to A/swine/Manitoba/01178/2006 and A/swine/Minnesota/01237/2006. Pairwise distance analysis of the challenge virus isolated on study day 32 from a vaccinated pig and a placebo pig showed that there were no nucleotide changes in the HA gene of the original virus isolate after pig passage.
Figure 1.
Phylogenetic tree for the hemagglutinin (HA) gene segments, based on nucleotide sequences of swine HA subtype 1 viruses (SwH1) found in the University of Minnesota Veterinary Diagnostic Laboratory and GenBank databases (GenBank accession numbers, when available, in parenthesis), along with the sequences of the challenge virus before and after inoculation (white diamonds) and the 2 H1N1 vaccine viruses (white circle and white triangle). The 4 clusters are SwH1γ (H1N2-like; black circles), SwH1β (reassortant H1N1-like; black squares), SwH1α (cH1N1 or classic-like; black diamonds), and SwH1δ (human-like H1; black triangles). There were 875 nucleotide positions in the final dataset. Bootstrap test results for similarity (500 replicates) are shown next to the branches.
Phylogenetic analysis also revealed that the nucleic acid sequence of the SwH1α challenge virus was only 87.2% similar to that of the SwH1γ vaccine virus. This difference between the HA genes was also demonstrated by the difference in number and positions of nucleotide changes within the putative antigenic sites (Table II): there were 11 changes in antigenic sites between the challenge virus and the SwH1γ vaccine virus and 24 changes between the challenge virus and the SwH1δ vaccine virus.
Table II.
Number and position of amino acid (AA) changes in putative antigenic sites for the hemagglutinin (HA) gene of the H1N1 vaccine strains compared with the challenge strain
| Antigenic site (total number of AA residues) | A/Sw/IA/110600/2000 | A/Sw/NC/031/2005 | ||
|---|---|---|---|---|
|
|
|
|||
| Number of changes | Position(s) | Number of changes | Position(s) | |
| Sa (13) | 4 | 125, 157, 161, 162 | 6 | 155, 157, 160, 161, 162, 164 |
| Sb (9) | 2 | 186, 190 | 8 | 153, 156, 186, 189, 190, 193, 194, 195 |
| Ca (14) | 4 | 137, 166, 170, 205 | 5 | 139, 142, 166, 170, 205 |
| Cb (8) | 1 | 73 | 5 | 69, 71, 72, 73, 74 |
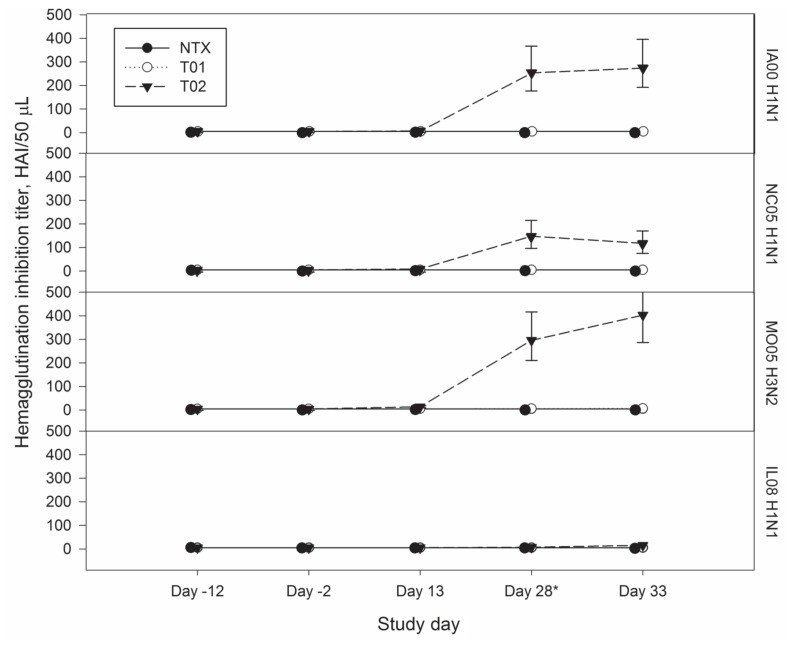
The reciprocal HI titers of antibody against the 4 viruses are shown for the 3 treatment groups in Figure 2. The titers were ≤ 10 (negative) against the vaccine and challenge viruses in all blood collections on days −12 and −2 and in the negative-control and placebo pigs on days 13 and 28. The vaccinated swine had titers 40 against the vaccine viruses and ≤ 20 against the challenge virus on days 28 and 33 (days 0 and 5 after challenge). For the SwH1γ vaccine virus, IA00 H1N1, there was no significant difference in titer between the treatment groups on days −2 and 13 (P = 1.0 and P = 0.1), but there was a significant difference on days 28 and 33 (P < 0.001). For the SwH1δ vaccine virus, NC05 H1N1, there was no significant difference in titer between the treatment groups on day −2 (P = 1.0), but there was a significant difference on days 13, 28, and 33 (P = 0.011, P < 0.001, and P < 0.001, respectively). For the SwH3 vaccine virus, MO05 H3N2, there was no significant difference in titer between the treatment groups on day −2 (P = 1.0), but there was a significant difference on days 13, 28, and 33 (P < 0.001). All the swine had a titer ≤ 20 against IL08 H1N1 on day 33 (5 d after challenge); there was no significant difference between the treatment groups on days −2 and 13 (P = 1.0), but there was a significant difference on days 28 and 33 (P < 0.001 and P < 0.001), the vaccinated animals having higher titers.
Figure 2.
Plots of back-transformed least-squares mean titers of hemagglutination inhibition (HI) antibodies against the 3 vaccine viruses (a, b, and c) and the challenge virus (d) from study day −12 to study day 33 for the negative-control pigs (NTX; black circles), placebo pigs (T01; white circles), and vaccinated pigs (T02; black triangles). IA00 H1N1 — A/swine/Iowa/110600/2000 (H1N1); NC05 H1N1 — A/swine/North Carolina/031/2005 (H1N1); MO05 H3N2 — A/swine/Missouri/069/2005 (H3N2); and IL08 H1N1 — A/swine/Illinois/02450/2008 (H1N1). The results for NTX have been staggered next to T01 for better visualization. The asterisk indicates the day of challenge.
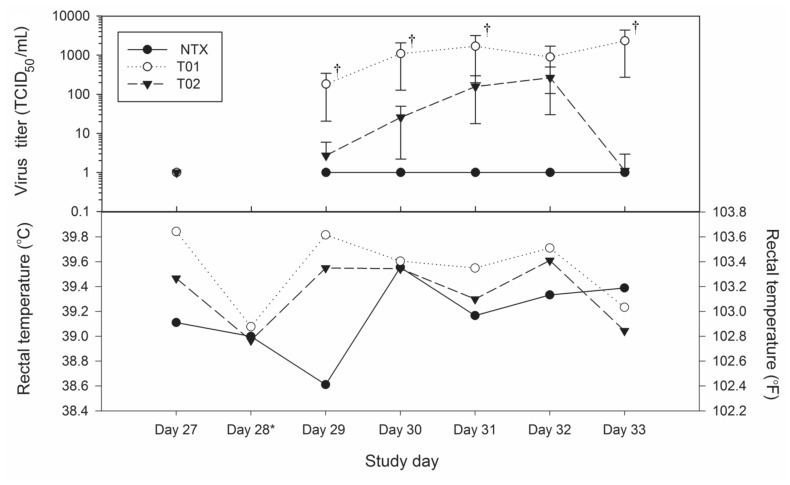
During the postchallenge period (days 29 to 33), mild clinical signs of respiratory disease were observed in 5 pigs (3 placebo and 2 vaccinated) for less than 24 h; 1 of the vaccinated pigs that was observed coughing also had labored breathing. Fever, defined by a rectal temperature 40.1°C, was observed for less than 48 h in 3 of the 9 pigs in the placebo group but none of the vaccinated pigs. Although the mean rectal temperature of the vaccinated pigs was consistently lower than that of the placebo group (Figure 3), the difference was not significant (P > 0.05).
Figure 3.
Plots of (a) back-transformed least-squares mean virus titers in the nasal swabs and (b) least-squares means of the pigs’ rectal temperatures. Virus titers of 0 are 1 on the logarithmic scale. The daggers indicate statistically significant differences in titer.
The nasal swabs collected from all animals 1 day before challenge were negative by real-time RT-PCR. The mean virus titer in the nasal swabs was significantly lower in the vaccinated pigs than in the placebo group at all time points except day 32 (4 d after challenge) (Figure 3). No virus was isolated from the nasal swabs of the vaccinated pigs 5 d after challenge. All of the nasal swabs that were negative by virus isolation were also negative by real-time RT-PCR (1 on the logarithmic scale in Figure 3). The geometric mean of the AUC of the virus titers, measured to examine nasal shedding during the 5-d study period, was significantly lower for the vaccinated pigs, at 491.1 TCID50/mL, than it was for the placebo pigs, at 13 922.9 TCID50/mL (P = 0.006).
The BALF and lung tissue samples from the placebo pigs were all positive by virus isolation, whereas the BALF and lung tissue samples from the vaccinated pigs were all negative. The geometric mean virus titer for the placebo pigs was 35 535.9 TCID50/mL in BALF and 1588.6 TCID50/mL in lung tissue, both significantly different from the titers for the vaccinated pigs (P < 0.001).
There were gross lesions in the lungs of all the pigs inoculated with the challenge virus. The lesions consisted of multifocal dense purple areas of lobular consolidation that occupied from 2% to 34% of the total lung volume. On average, more than half of the lung lesions observed were in the right and left middle lobes. The back-transformed least-squares means of the proportion of lung tissue consolidated was 12% [confidence interval (CI) 4% to 24%] for the placebo group and 9% (CI 2% to 20%) for the vaccinated group; the difference was not significant (P = 0.403). The NTX pigs (2 sentinels and 2 negative-controls) had no macroscopic lung lesions.
Microscopic lung lesions
The acute lesions of purulent bronchiolitis that were observed in the epithelial lining of large, medium, and small bronchioles were accompanied by loss of apical cilia, epithelial cell necrosis with intraluminal sloughing and attenuation, areas of epithelial hyperplasia, and transmigration of large numbers of neutrophils admixed with a few lymphocytes and macrophages. There was rare bronchiolitis obliterans in the most severely affected lung sections. Multifocally, there was peribronchiolar to lobular pneumonia characterized by moderate to large numbers of neutrophils, lymphocytes, and macrophages within the alveolar spaces and occasionally expanding the alveolar septa. Five of the nine vaccinated pigs had a lesion score of 1, with only a few isolated airways affected, compared with just 1 placebo pig. Three of the nine placebo pigs had the most severely affected lung sections, with a lesion score of 4. There was no significant difference between the categorized microscopic lung scores of the placebo group and those of the vaccinated group (P = 0.14). The NTX pigs had no microscopic lung lesions.
Additional diagnostic testing
The PCR tests for NAEU variants of PRRSV and M. hyopneumoniae and the aerobic cultures for bacterial pathogens all had negative results for all the pigs. The PCV-2 PCR tests had negative results for all of the pigs except for 1 pig in the placebo group and 2 pigs in the vaccinated group. The cycle threshold values for these results were 21.88, 16.88, and 35.35, respectively.
Discussion
Vaccination against influenza is an important part of a comprehensive plan to limit the effects of IAV infection in a herd by reducing the amount of clinical disease and virus shedding (1,22) and thus mitigate the related economic losses. These losses can be significant, particularly in growing pigs, and are due to increased mortality, decreased feed efficiency, and the costs of treating secondary bacterial infections (1).
The rapid evolution of influenza viruses over the last decade in North America, particularly those of the SwH1 subtype, has resulted in remarkable genetic and antigenic variability. Since the efficacy of a vaccine is directly linked to genetic and antigenic matching of the vaccine virus to the challenge virus, the variability in circulating field strains is a major challenge for the North American swine industry. The commercial vaccine used in this study, containing SwH1γ and SwH1δ viruses, has demonstrated cross-protection against challenge with the SwH1γ virus A/swine/Minnesota/000581/2001 (H1N1) and the SwH1β virus PAH-NADC11 (H1N1), with 90.1% and 80.5% reductions, respectively, in gross lung lesions (23). Other efficacy studies of this vaccine have also demonstrated cross-reactive HI antibodies and significant reductions in gross lung lesions after challenge with classic swine H1N1 (cH1N1) virus A/swine/Iowa/40776/1992 and SwH1β virus A/swine/Minnesota/00040/2002 (H1N1) (24,25). In the most recent studies involving the 2009 pandemic virus (pH1N1) A/California/04/2009 (H1N1), which contains an SwH1γ HA gene, the vaccine-induced HI antibody titers of very low cross-reactivity, yet the vaccinated animals had significant reductions in gross lung lesions and nasal-swab virus titers (26).
The challenge viruses in these studies were from early in the evolution of triple-reassortant swine viruses (1999 to 2002), except for A/swine/Iowa/40776/1992, a cH1N1 virus from before the introduction of the triple-reassortant internal gene (TRIG) cassette (3). Since the viruses currently circulating in Canada with the cH1N1-like HA gene have the TRIG cassette (7), the present study was conducted to evaluate the efficacy of the commercial vaccine in pigs challenged with a contemporary SwH1α field isolate of Canadian swine origin that had classic-like swine H1 and N1 genes and the TRIG cassette.
The challenge virus, IL08 H1N1, was determined by phylogenetic analysis of the HA gene sequence to be an α-cluster field isolate that was more genetically related (with > 99% nucleotide similarity) to isolates from Manitoba (in the UMVDL sequence database) than to influenza viruses previously isolated from Illinois. Further investigation revealed that the farm in Illinois where the isolate originated was surrounded by grower/finisher sites that had been recently populated with swine imported from Canada. Since none of the viruses isolated from this site or other sites within this multisite production system are closely related to IL08 H1N1 by phylogenetic analysis, and although there was no virus available for comparison from the imported pigs, it is most likely that IL08 H1N1 is a virus of Canadian swine origin.
Vaccine-induced protection against influenza in the best-case scenario should result in a reduction of gross and histopathological lesions in the lungs, clinical signs, and frequency of virus isolation in nasal swabs and BALF. The postmortem examinations conducted on day 33 revealed that all challenged pigs, both vaccinated and placebo, had gross lesions in the lungs; there was no significant difference between the groups in the proportion of lung tissue consolidated (P = 0.403). Two-thirds of the placebo pigs had gross lesions in 10% or more of the lung, compared with only one-third of the vaccinated pigs. There was also no significant difference between the vaccinated and placebo groups in the categorized microscopic lung scores (P = 0.14). However, the moderate to severe lesions (scored 3–4) were found in the lungs of 78% of the placebo pigs compared with only 22% of the vaccinated pigs.
All of the vaccinated pigs had a reciprocal HI titer 40 against the vaccine viruses, a titer that is considered protective (27). However, the reciprocal HI titers against the challenge virus were ≤ 10 at the time of challenge, reflecting the HA sequence differences between the vaccine virus and the α-cluster SwH1N1 virus. Although virus was detected and lung lesions were present in almost all of the challenged pigs after challenge, the infection induced only mild clinical signs that included fever lasting 24 to 48 h in a few pigs and rare coughing. The virus titers in the nasal swabs taken during the challenge period (days 29 to 33) showed a reduction in virus shedding for the vaccinated swine compared with the placebo group after challenge, and no virus was detected in the vaccinated pigs on day 5 after challenge. There was also a significant difference between treatment groups in the geometric mean virus titer for both BALF and lung tissue (P < 0.001), with no virus being detected in the BALF or lung tissue collected from vaccinates on day 5 after challenge. Thus, although the vaccine did not reduce lung lesions under the conditions of this study, it did reduce replication of the virus in the lung and nasal shedding.
In conclusion, the results indicate that vaccination with Flusure XP reduced the amount of virus in nasal secretions, lungs, and BALF in swine experimentally challenged with an SwH1α cluster virus that is genetically similar to viruses currently circulating in Canadian swine. This indicates that the vaccine helped to protect against challenge with a virus from an SwH1 cluster not represented in the vaccine.
Acknowledgments
The funding for this project was provided by Pfizer Animal Health, Kalamazoo, Michigan, USA, under study report 3121R-02-09-789. The authors thank Al Ducommun, Jackie Gaulke, Michele Leiferman, Elissa Schlueter, Paul Syverson, Ling Tong, Evan Van Beusekom, and Wendy Wiese for their technical expertise and contributions to this work. The authors also thank Drs. Matt Allerson, Seth Baker, Cesar Corzo, Alejandrina de Silva, Daniel Linhares, Devi Patnayak, and Genevieve Remmers for their assistance with this project.
References
- 1.Torremorell M, Juarez A, Chavez E, Yescas J, Doporto JM, Gramer M. Procedures to eliminate H3N2 swine influenza virus from a pig herd. Vet Rec. 2009;165:74–77. doi: 10.1136/vetrec.165.3.74. [DOI] [PubMed] [Google Scholar]
- 2.Kitikoon P, Strait EL, Thacker EL. The antibody responses to swine influenza virus (SIV) recombinant matrix 1 (rM1), matrix 2 (M2), and hemagglutinin (HA) proteins in pigs with different SIV exposure. Vet Microbiol. 2008;126:51–62. doi: 10.1016/j.vetmic.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 3.Vincent AL, Lager KM, Ma W, et al. Evaluation of hemagglutinin subtype 1 swine influenza viruses from the United States. Vet Microbiol. 2006;118:212–222. doi: 10.1016/j.vetmic.2006.07.017. [DOI] [PubMed] [Google Scholar]
- 4.Vincent AL, Ma W, Lager KM, Gramer MR, Richt JA, Janke BH. Characterization of a newly emerged genetic cluster of H1N1 and H1N2 swine influenza virus in the United States. Virus Genes. 2009;39:176–185. doi: 10.1007/s11262-009-0386-6. [DOI] [PubMed] [Google Scholar]
- 5.Karasin AI, Carman S, Olsen CW. Identification of human H1N2 and human-swine reassortant H1N2 and H1N1 IAVs among pigs in Ontario, Canada (2003 to 2005) J Clin Microbiol. 2006;44:1123–1126. doi: 10.1128/JCM.44.3.1123-1126.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lorusso A, Vincent AL, Harland ML, et al. Genetic and antigenic characterization of H1 influenza viruses from United States swine from 2008. J Gen Virol. 2011;92:919–930. doi: 10.1099/vir.0.027557-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nfon CK, Berhane Y, Hisanaga T, et al. Characterization of H1N1 swine influenza viruses circulating in Canadian pigs in 2009. J Virol. 2011;85:8667–8679. doi: 10.1128/JVI.00801-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR. Universal primer set for the full-length amplification of all influenza A viruses. Arch Virol. 2001;146:2275–2289. doi: 10.1007/s007050170002. [DOI] [PubMed] [Google Scholar]
- 9.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 10.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 11.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 12.Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 13.Caton AJ, Brownlee GG, Yewdell JW, Gerhard W. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype) Cell. 1982;31:417–427. doi: 10.1016/0092-8674(82)90135-0. [DOI] [PubMed] [Google Scholar]
- 14.Qi X, Lu CP. Genetic characterization of novel reassortant H1N2 influenza A viruses isolated from pigs in southeastern China. Arch Virol. 2006;151:2289–2299. doi: 10.1007/s00705-006-0796-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meguro H, Bryant JD, Torrence AE, Wright PF. Canine kidney cell line for isolation of respiratory viruses. J Clin Microbiol. 1979;9:175–179. doi: 10.1128/jcm.9.2.175-179.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Villegas P. Titration of biological suspensions. In: Dufour-Zavala L, Swayne DE, Glisson JR, editors. A Laboratory Manual for the Isolation, Identification, and Characterization of Avian Pathogens. 5th ed. Athens, Georgia: American Association of Avian Pathologists; 2008. pp. 218–221. [Google Scholar]
- 17.Pedersen JC. Hemagglutination-inhibition test for avian influenza virus subtype identification and the detection and quantitation of serum antibodies to the avian influenza virus. In: Spackman E, editor. Methods in Molecular Biology, Vol 436: Avian Influenza Virus. Totowa, New Jersey: Humana; 2008. pp. 53–66. [DOI] [PubMed] [Google Scholar]
- 18.Halbur PG, Paul PS, Frey ML, et al. Comparison of the pathogenicity of two US porcine reproductive and respiratory syndrome virus isolates with that of the Lelystad virus. Vet Pathol. 1995;32:648–660. doi: 10.1177/030098589503200606. [DOI] [PubMed] [Google Scholar]
- 19.Richt JA, Lager KM, Janke BH, Woods RD, Webster RG, Webby RJ. Pathogenic and antigenic properties of phylogenetically distinct reassortant H3N2 swine influenza viruses cocirculating in the United States. J Clin Microbiol. 2003;41:3198–3205. doi: 10.1128/JCM.41.7.3198-3205.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spackman E, Senne DA, Myers TJ, et al. Development of a real-time reverse transcriptase PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes. J Clin Microbiol. 2002;40:3256–3260. doi: 10.1128/JCM.40.9.3256-3260.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spackman E, Suarez DL. Type A influenza virus detection and quantitation by real-time RT-PCR. Methods Mol Biol. 2008;436:19–26. doi: 10.1007/978-1-59745-279-3_4. [DOI] [PubMed] [Google Scholar]
- 22.Olsen CW, Brown IH, Easterday BC, Van Reeth K. Diseases of Swine. Ames, Iowa: Blackwell; 2006. Swine influenza; pp. 469–482. [Google Scholar]
- 23.Rapp-Gabrielson VJ, Wicklund E, Ficken M. Efficacy of Flusure® against challenge with heterologous reassortant swine influenza H1N1 viruses. Proceedings of the Allen D. Leman Swine Conference; 2005 Sept 17–20; St. Paul, Minnesota. p. 14. [Google Scholar]
- 24.Kitikoon P, Nilubol D, Erickson BJ, et al. The immune response and maternal antibody interference to a heterologous H1N1 swine influenza virus infection following vaccination. Vet Immunol Immunopathol. 2006;112:117–128. doi: 10.1016/j.vetimm.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 25.Gramer MR, Rossow KD. Epidemiology of swine influenza and implications of reassortment. Proceedings of the Allen D. Leman Swine Conference; 2004 Sept 18–21; St. Paul, Minnesota. 2004. pp. 69–73. [Google Scholar]
- 26.Vincent AL, Ciacci-Zanella JR, Lorusso A, et al. Efficacy of inactivated swine influenza virus vaccines against the 2009 A/H1N1 influenza virus in pigs. Vaccine. 2010;28:2782–2787. doi: 10.1016/j.vaccine.2010.01.049. [DOI] [PubMed] [Google Scholar]
- 27.Hancock K, Veguilla V, Lu X, et al. Cross-reactive antibody responses to the 2009 pandemic H1N1 influenza virus. N Engl J Med. 2009;361:1945–1952. doi: 10.1056/NEJMoa0906453. [DOI] [PubMed] [Google Scholar]