Abstract
Non-long terminal repeat retroelements continue to impact the human genome through cis-activity of long interspersed element-1 (LINE-1 or L1) and trans-mobilization of Alu. Current activity is dominated by modern subfamilies of these elements, leaving behind an evolutionary graveyard of extinct Alu and L1 subfamilies. Because Alu is a nonautonomous element that relies on L1 to retrotranspose, there is the possibility that competition between these elements has driven selection and antagonistic coevolution between Alu and L1. Through analysis of synonymous versus nonsynonymous codon evolution across L1 subfamilies, we find that the C-terminal ORF2 cys domain experienced a dramatic increase in amino acid substitution rate in the transition from L1PA5 to L1PA4 subfamilies. This observation coincides with the previously reported rapid evolution of ORF1 during the same transition period. Ancestral Alu sequences have been previously reconstructed, as their short size and ubiquity have made it relatively easy to retrieve consensus sequences from the human genome. In contrast, creating constructs of extinct L1 copies is a more laborious task. Here, we report our efforts to recreate and evaluate the retrotransposition capabilities of two ancestral L1 elements, L1PA4 and L1PA8 that were active ∼18 and ∼40 Ma, respectively. Relative to the modern L1PA1 subfamily, we find that both elements are similarly active in a cell culture retrotransposition assay in HeLa, and both are able to efficiently trans-mobilize Alu elements from several subfamilies. Although we observe some variation in Alu subfamily retrotransposition efficiency, any coevolution that may have occurred between LINEs and SINEs is not evident from these data. Population dynamics and stochastic variation in the number of active source elements likely play an important role in individual LINE or SINE subfamily amplification. If coevolution also contributes to changing retrotransposition rates and the progression of subfamilies, cell factors are likely to play an important mediating role in changing LINE-SINE interactions over evolutionary time.
Keywords: Alu, extinction, LINE-1, L1, ORF1 protein, ORF2 protein, retroelement, SINE amplification
Introduction
Long interspersed element-1 (LINE-1 or L1) is the dominant human non-long terminal repeat autonomous retroelement and has been active in mammalian genomes for more than 170 My (Smit 1999). The human genome has been significantly impacted by the activity of L1, through both self-mobilization and trans-mobilization of the SINE Alu. Together, L1 and Alu repeat sequences account for at least a third of the human genome (Lander et al. 2001), and more recent analyses suggest this may be a gross underestimation (de Koning et al. 2011). Following retrotransposition, the active Alu and L1 copies lose functionality as they accumulate mutations at a neutral rate, leaving older copies with higher sequence degradation than newer copies. Phylogenetic analysis of L1 families has shown that L1 subfamilies follow a linear pattern, whereby a single L1 lineage proliferates, differentiates, and is eventually replaced by a new dominant subfamily (Deininger et al. 1992; Smit et al. 1995; Boissinot and Furano 2001). Alu subfamilies follow a similar pattern with a progression of dominant subfamilies over the course of primate evolution (Shen et al. 1991).
L1 elements contain two open reading frames (ORF1 and ORF2) that code for proteins essential for L1 retrotransposition. Trans-mobilization of short interspersed elements (SINEs), by contrast, is only ORF2 dependent (Dewannieux et al. 2003; Wallace et al. 2008). Because Alu requires L1 to retrotranspose, it is conceivable that competition between these retroelements has triggered antagonistic coevolution between LINEs and SINEs and altered their interactions over evolutionary time. In some cases, one or both elements could be driven to extinction within a lineage. For example, coextinction of L2 and its proposed SINE partner, MIR, has been observed in humans (Lander et al. 2001). In another example, sigmodontine rodents lost both functional L1 and B1 SINEs (Rinehart et al. 2005). In this case, B1 silencing appears to have preceded L1 extinction. Thus, the mere presence of an active L1 is not necessarily sufficient to support SINE activity, suggesting that host factors and/or changes within the SINE itself could affect retrotranspositional capability.
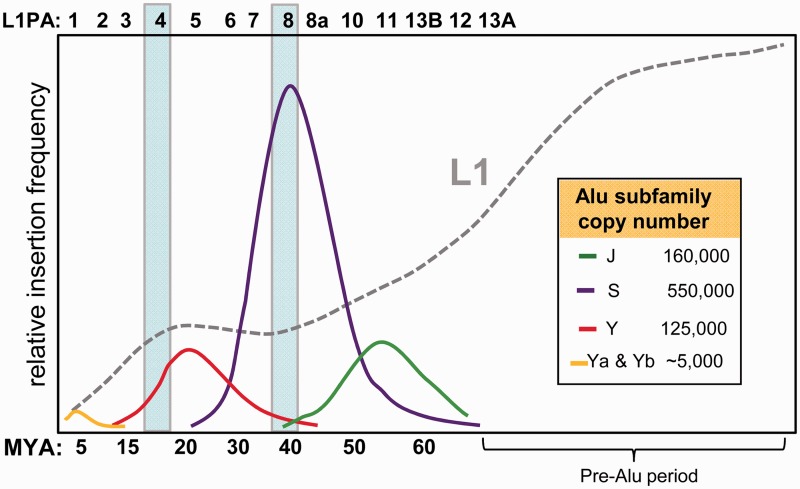
Several studies have used retroelement insertion sequence divergence and/or presence/absence data from primate genomes (Shen et al. 1991; Ohshima et al. 2003; Khan et al. 2006; Bennett et al. 2008) to evaluate the temporal proliferation of mammalian retroelements. We created a simplified schematic of some of these findings in figure 1 by showing the amplification history of the major Alu subfamilies relative to that of L1. L1 went through a long period of high amplification, followed by a steady decline in activity from approximately 65 Ma to the current relatively low rate. Evaluation of Alu amplification reveals that different Alu subfamilies experienced peak activity during discrete periods, with waves of Alu subfamily activity occurring at ∼15–20 Ma, ∼40–50 Ma, and ∼55 Ma with the proliferation of Alu Y, S, and J subfamilies, respectively. The relatively less abundant young Alu Ya and Yb subfamilies are currently active and most likely account for all human-specific insertions (Batzer and Deininger 2002; Hedges et al. 2004). The decline in L1 activity roughly coincides with the initial emergence of Alu elements (∼65 Ma). Ohshima et al. (2003) compared the evolutionary proliferation of Alu and L1 repeats in humans, as well as processed pseudogenes, and showed that peak Alu and pseudogene amplification occurred simultaneously at approximately 40–50 Ma. This observation led to the suggestion that the dominant ancestral L1 subfamilies of the era might have mobilized RNAs in trans at accelerated rates relative to other L1 subfamilies (Ohshima et al. 2003). During this peak period of amplification, the Alu J and Alu S subfamilies were actively generating the majority (∼80% of the 1.1 million) of Alu copies in the human genome. Interestingly, the period of elevated Alu Y subfamily amplification (15–20 Ma) also coincides with the emergence of another L1-dependent nonautonomous element, SINE/VNTR/Alu (SVA), in the hominid lineage (Wang et al. 2005).
Fig. 1.
Age distribution of Alu and L1 subfamilies. This schematic depicts the rate of insertion for Alu and L1 elements over evolutionary time. The relative insertion frequency for L1 (all subfamilies combined) is represented by the gray dotted line and is set at a scale 4× larger relative to Alu to emphasize the changing rate of amplification coinciding with the emergence of the indicated Alu subfamilies. The corresponding active subfamilies from the L1PA family at each time period are indicated above. Individual Alu subfamilies are shown with their total copy number indicated in the inset. The two timeframes that include peak activity periods for L1PA4 (∼18 Ma) and L1PA8 (∼40 Ma) are indicated by shaded boxes. Data were adapted from Shen et al. (1991), Ohshima et al. (2003), Khan et al. (2006), and Bennett et al. (2008).
Here, we present data that demonstrate the reconstruction of functional full-length L1 elements from two extinct human L1 subfamilies that were active during periods of increased Alu amplification or rapid L1 protein evolution. We show that they are retrocompetent in an ex vivo tissue culture assay, both for L1 cis-mobilization and trans-mobilization of Alu. We find limited evidence of differential associations between Alu and L1 subfamilies, suggesting that other factors are likely the primary mediators of their changing interactions over evolutionary time.
Materials and Methods
Constructs
A schematic of the basic Alu- and L1-tagged vectors is shown in figure 2. The “SINE”-neoTET constructs (pAluY-neoTET, pAluSg1-neoTET, pAluSx-neoTET, and pAluJo-neoTET) were created by substituting the Ya5 Alu element from pAluYa5-neoTET (Kroutter et al. 2009) with the different Alu subfamily consensus sequences using a BamHI site (5′ of 7SL promoter enhancer sequence) and the introduced AatII site (fig. 2). The AluSx consensus sequence (previously known as AluPS) differs at position 225 (G instead of C) (Alemán et al. 2000).
Fig. 2.
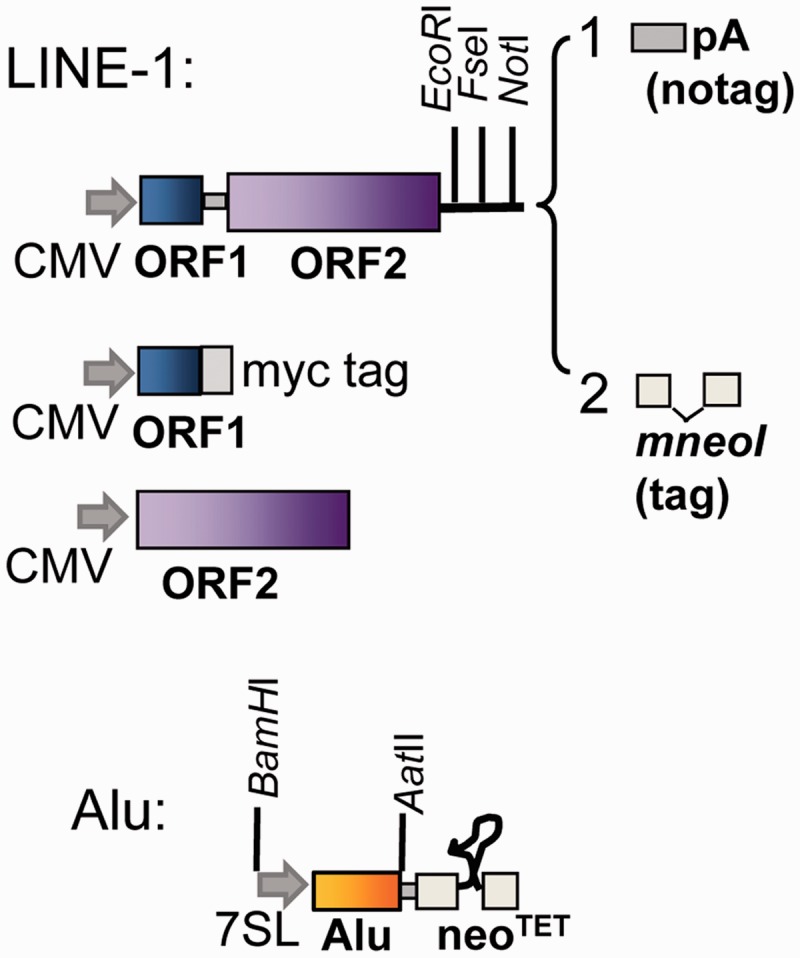
Schematic of the L1 and Alu constructs. A representation of the basic components of the constructs is shown. The L1 constructs contain the codon-optimized ORF1 and ORF2 separated by the L1RP inter-ORF sequence (Wagstaff et al. 2011) or the wild-type sequence of the L1PA8 ORFs. Two types of L1 constructs were built, which differ only at their 3′-end: 1, untagged containing the SV40 polyadenylation signal (pA) and 2, tagged with the neomycin indicator cassette designed for retrotransposition assays (mneoI). The individual ORF1 (blue) and ORF2 (purple) sequences were cloned downstream of the CMV promoter of the expression vector pBudCE4.1 (Invitrogen). The ORF1 was cloned, so that the protein will contain a myc-his tag (myc) at the carboxy terminus. The Alu subfamily (yellow) constructs are tagged with the neomycin indicator cassette (neoTET) designed for retrotransposition assays. RNA transcription is performed by the CMV promoter (L1) or the internal pol III promoter of the Alu enhanced by the upstream sequence of the 7SL gene (shown as gray arrows). The retrotransposition indicator cassettes (mneoI or neoTET) contain an inverted neomycin resistance gene disrupted by an intron that will splice only from a transcript generated by the CMV or Alu promoter. The neoTET contains a modification of the ribozyme from Tetrahymena (represented as a looped line) that functions as a self-splicing intron (Esnault et al. 2002). Only retrotransposed copies of the spliced RNA will confer G418 resistance. Some of the unique restriction sites used in the construction of the vectors are shown.
JM101/L1.3, referred to as “wild type” L1, contains a full-length copy of the L1.3 element tagged with the mneoI indicator cassette cloned in pCEP4 (Invitrogen) (Dombroski et al. 1993; Sassaman et al. 1997).
Reconstruction of Extinct L1 Elements
The codon-optimized L1 PA4 and PA8 and wild-type L1PA8 ORF1 and ORF2 consensus sequences were synthesized by Blue Heron Biotechnology, Inc (Bothell, WA) or GenScript. Codon optimization of the sequences was performed using Primo Optimum 3.4 (http://www.changbioscience.com/primo/primoo.html). Note: the L1PA8 constructs contain the corrected version of the consensus sequence (table 2). All bicistronic L1 constructs were built using pBS-L1PA1CHmneo as base (Wagstaff et al. 2011) by substituting the L1 PA1 ORF1 and ORF2 coding sequences with the corresponding synthesized L1 sequences. Different cassettes were added at the 3′-end of each L1 subfamily construct (fig. 2):
pBS-L1PA1CHmneo, pBS-L1PA4CHmneo, and pBS-L1PA8CHmneo, referred to as the “tagged” vectors, contain the codon-optimized ORF1 and ORF2 of the consensus sequence of each subfamily and the mneoI cassette including the SV40 polyadenylation signal (pA) from JM101/L1.3 (Dombroski et al. 1993).
pBS-L1PA8WTmneo contains the corrected version of the “wild type” consensus sequence of L1PA8 (Khan et al. 2006), with the 11 modified codons as described in table 2.
pBS-L1PA1CHnotag, pBS-L1PA4CHnotag, and pBS-L1PA8CHnotag, referred to as the “no tag” constructs, contain an SV40 pA at the 3′-end that was introduced into the EcoRI-FseI sites (fig. 2).
Table 2.
Analysis of Codon Changes Involved in the Modified L1PA8 Consensus Sequence.
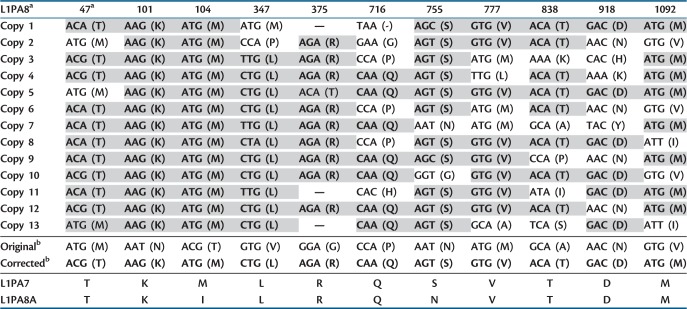 |
Note.–Residues matching the corrected sequence are in bold and highlighted gray.
aAmino acid sequence numbers using the ORF2 L1RP sequence as reference. Codons are indicated with amino acid in parentheses. Absent codons (deletions) are designated by dashes.
bThe original consensus, corrected, derived subfamily, and ancestral subfamily are shown at the bottom for the indicated codon positions.
The individual ORFs of the different L1 elements were all cloned into the expression vector pBudCE4.1 (Invitrogen), under control of the cytomegalovirus (CMV) promoter:
pBudORF2CH (Wagstaff et al. 2011) and pBudORF1opt (Wallace et al. 2008) were created using the codon-optimized L1RP as a source for the ORF2 and ORF1 coding sequences. These constructs are used for the expression of the L1PA1 ORF1 and ORF2.
pBudORF1PA1CH-myc, pBudORF1PA4CH-myc, and pBudORF1PA8CH-myc were generated by cloning the polymerase chain reaction (PCR)-amplified codon-optimized consensus sequences of each ORF into the HindIII-BamHI sites of the pBudCE4.1 vector in a manner that removes the stop codon of the ORF1, so the expressed protein will contain the myc-his tag at the carboxy terminus (fig. 2). The following primers were used in the amplification of ORF1: 5-AGACCCAAGCTTAGCTAAAACCACAAAGATG-3′ and 5-TGTTCGGATCCGATCTTGGTGTGCTTCTGCAGGGG-3′ for ORF1PA8 or 5-TGTTCGGATCCCATCTTGGCGTGGTTTTGCAGGGG-3′ for ORF1 PA1 and ORF1PA4.
pBudORF2PA4CH and pBudORF2PA8CH were generated by cloning the codon-optimized consensus sequences of each ORF that retain the stop codon into the HindIII-BamHI sites of the pBudCE4.1 vector (fig. 2).
Plasmids were independently purified in triplicate by either alkaline lysis and twice purified by cesium chloride buoyant density centrifugation or by using the QIAGEN Plasmid Plus Maxi kit, following the manufacturer’s protocol. DNA quality was also evaluated by the visual assessment of ethidium bromide-stained agarose gel-electrophoresed aliquots to evaluate purity and quality. All new constructs were sequence verified.
Analysis of Nonsynonymous and Synonymous Substitutions across L1 Subfamilies
Consensus sequences for the analysis were from Khan et al. (2006) but with the 11 modified codons of L1PA8 ORF2 as detailed in table 2. Domain breakpoints for the ORF2 protein were determined as follows, with residue numbers corresponding to L1PA1 ORF2p: the N terminus endonuclease included residues 1–239, in accordance with the well-established domain (Feng et al. 1996; Cost et al. 2002; Weichenrieder et al. 2004); the reverse transcriptase (RT) domain included residues 511–773, following the boundaries as defined by the Conserved Domain Database (CDD v3.05-42589 PSSMs [Marchler-Bauer et al. 2005]); and the remaining “inter endo RT” (residues 240–510) and “cys” (residues 774–1,275) domains were simply the remaining regions of ORF2p 5′ of the RT and 3′ of the RT, respectively. Nonsynonymous and synonymous substitution rates between temporally adjacent L1 subfamilies were computed using DnaSP v5 (Librado and Rozas 2009).
LINE and SINE Assays
Transient L1 or Alu retrotransposition assays were performed as described previously with some minor modifications (Kroutter et al. 2009). Briefly, HeLa cells (ATCC CCL2) were seeded in T25 or T75 flasks at a density of 2 × 105 or 5 × 105 cells, respectively. Transient transfections were performed the following day using Lipofectamine Plus (InVitrogen) following the manufacturer’s protocol. L1 retrotransposition was assayed in T25 flasks by transfecting cells with 0.4 µg of the L1 constructs. To evaluate Alu retrotransposition, cells were seeded in six-well plates at a density of 1.0 × 105 cells per well. The cells were transfected with 1 µg of the ORF2 expression vector or with 1 µg of the untagged L1 subfamily construct and varying amounts of the tagged Alu subfamily constructs (0.1–1 µg) as indicated. Empty vector was used in the mix to equalize the amount of total DNA used in each transfection reaction. The following day, the cells were treated with the appropriate selection media containing 400 µg/ml Geneticin/G418 (Fisher Scientific). After 14 days, cells were fixed and stained for 30 min with crystal violet (0.2% crystal violet in 5% acetic acid and 2.5% isopropanol). For transfections using the RT inhibitor 2′,3′-didehydro-3′-deoxy-thymidine (d4t; Sigma-Aldrich), a final concentration of 50 µM d4t was added to the media at the time of transfection and maintained with subsequent media changes for a period of 7 days.
PCR Evaluation of L1 Inserts
Colonies of G418 resistant cells were pooled, and DNA was extracted using the DNA-Easy kit (Qiagen) following the manufacturer’s recommended protocol. PCR was performed for 35 cycles at 58°C annealing temperature with a 1 min extension using Taq polymerase with the following primers designed to flank the intron disrupting the neomycin gene (fig. 4B): RNeo-Exon1: 5′-ATGGGATCGGCCATTGAACAAGATG-3′ and FNeo-Exon2: 5′-GCAAGGTGAGATGACAGGAGATCC-3′. Amplification products containing the unspliced intron are expected to be 1,233 bp, whereas spliced products with the intron removed are 330 bp.
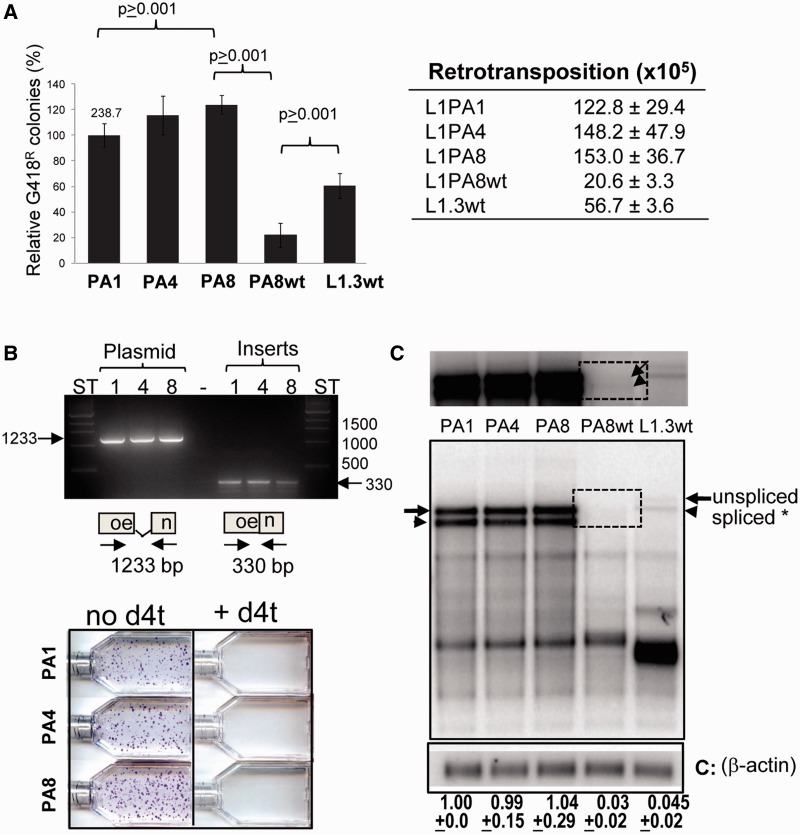
Fig. 4.
The reconstructed L1PA4 and L1PA8 are retrotransposition competent. (A) Relative retrotransposition efficiencies of reconstructed L1 elements in HeLa cells. The retrotransposition capability of the individual tagged L1 constructs: codon-optimized L1PA1 (PA1), L1PA4 (PA4), and L1PA8 (PA8) and the wild-type L1PA8 (PA8wt) and JM101/L1.3 (L1.3 wt) are shown. Columns represent the G418R colony means normalized relative to the L1PA1 with the standard deviation shown as error bars. The mean number of G418R before normalization for the L1PA1 is shown above the column. Results from Student’s paired t-test are indicated (n ≥ 4). The panel on the right shows the mean ± standard error of the mean (SEM) of G418 resistant colonies observed for each L1 construct evaluated (n ≥ 4). (B) Verification of L1PA4 and L1PA8 retrotransposition events. Top panel shows the PCR analysis of HeLa cells that were transfected with the tagged L1PA1, L1PA4, and L1PA8 vectors. PCR analysis was performed using primers designed to anneal to the sequence flanking the intron disrupting the neomycin gene (neo). Plasmid DNA from each L1 construct was used as unspliced control (left). The annealing locations of the primers are shown in the schematic of the neo cassette plus intron (1,233 bp) and without intron (330 bp). DNA from the tagged L1 plasmids: L1PA1 (1), L1PA4 (4), and L1PA8 (8) was used as control for the unspliced cassette. Results from DNA extracts from pooled G418R colonies generated by the indicated L1 constructs are shown as "Inserts." ST, size standard lanes are indicated. The lower panel shows a representative L1PA4 and L1PA8 retrotransposition experiment in the presence or absence of the RT inhibitor d4t. (C) Evaluation of the RNA profiles of the reconstructed L1 constructs. HeLa cells were transiently transfected with the tagged optimized L1PA1CH (PA1), L1PA4CH (PA4), L1PA8CH (PA8), and “wild type” L1PA8wt (PA8wt) and L1.3 construct JM101/L1.3 (L1.3 wt). Poly-A selected RNA was hybridized with a strand-specific riboprobe to the neomycin resistance gene or to beta-actin (bottom panel indicated by C). The full-length unspliced tagged L1 transcript (arrow) and the full-length transcript with spliced neo tag (arrowhead) are indicated. The top panel shows a longer exposure of the blot. The dotted box highlights the location of the weak signal of the L1 PA8 wild-type transcripts for easier visualization. The faster migrating bands are likely common splice transcript variants previously shown to be generated by L1 elements (Belancio et al. 2006; Wagstaff et al. 2011). The spliced full-length L1transcript was normalized to beta-actin and calculated relative to the L1PA1 construct (designated as 1.0). The mean ± SEM for the quantification results for each construct is indicated below (n = 3). No significant differences were observed between the optimized L1PA1, L1PA4, and L1PA8 spliced transcripts (one-way analysis of variance, F = 0.80, P = 0.923604; n = 5). RNA levels for L1PA8WT and L1.3 are significantly lower than their optimized L1 counterparts (paired t-test, P < 0.0001; n = 3).
Northern Blot Analysis
Cells were harvested 24 h post-transfection. RNA extraction and poly(A) selection were performed as described previously (Perepelitsa-Belancio and Deininger 2003). The polyadenylated RNA species were evaluated in a 2% (Alu and ORF1 constructs) or a 1% (L1 and ORF2 constructs) agarose-formaldehyde gel and transferred to a Hybond-N nylon membrane (Amersham Biosciences). The RNA was UV cross-linked to the membrane using ultraviolet (UV) light (GS Gene linker, BioRad). The membrane was preincubated in hybridization solution: 30% formamide, 1X Denhardt’s solution, 1% SDS, 1 M NaCl, 100 µg/ml salmon sperm DNA, and 100 µg/ml yeast t-RNA at 60°C for at least 3 h. The DNA templates containing the T7 promoter for riboprobe generation were generated by PCR amplification. For the 3′-region of the neomycin gene used primers: T7neo(-): 5′-TAATACGACTCACTATAAGGACGAGGCAGCG-3′ and Neo northern(+): 5′-GAAGAACTCGTCAAGAAGG-3′; for the ORF2 used primers: T7ORF2CH180 5′-TAATACGACTCACTATAGGCTGGATGCCCTTGATCTCC-3′ and F-ORF2CH180 5′-AAGATCATCCGGGCCATCTACGA-3′; and for the myc-his tag 3′-region of the tagged ORF1 used primers: T7mychis: 5′-TAATACGACTCACTATAGGGATGTCT-3′ and F-mychis: 5′-TGGTGATGGTGATGATGCATCTTGGC-3′. We used a commercially available construct to generate the riboprobe for β-actin (Ambion). Riboprobes were generated by incorporating 32P-CTP (Amersham Biosciences) label using the MAXIscript T7 kit (Ambion) following the manufacturer’s recommended protocol. The radiolabeled probes were purified by filtration through a NucAway Spin column (Ambion). Separate hybridizations were performed overnight with 4–12 × 106 cpm/ml of each individual probe at 60°C. The membrane was washed twice at high stringency (0.1× Ssline-sodium citrate [SSC], 0.1% sodium dodecyl sulfate [SDS]) at 60°C before analysis using a Typhoon Phosphorimager (Amersham Biosciences) and the ImageQuant software.
Western Blot Analysis
Two to four T75 flasks of HeLa cells (4 × 106/flask) were transiently transfected with 6 µg of plasmid per T75. Cells were harvested 24 h post-transfection. Equal amount of protein extracts were electrophoresed on 3–8% Tris-acetate gel (Invitrogen). Proteins were transferred to a nitrocellulose membrane using the iBlot gel transfer system using the manufacturer’s recommended settings (Invitrogen). Blots were blocked overnight in phosphate buffer saline (PBS) pH 7.4, 0.05% Tween 20, 5% nonfat dry milk (Biorad) at 4°C. A mouse monoclonal anti-myc (clone 9E10, Upstate) was used to detect the myc-tagged ORF1p. Antibodies against β-actin and secondary horse radish peroxidase (HRP)-conjugated antibodies were purchased from Santa Cruz Biotechnology Inc. The membrane was incubated for 1 h at room temperature with the primary or secondary antibody diluted 1:500 and 1:5,000 in PBS pH 7.4, 0.05% Tween 20, 3% nonfat dry milk (Biorad), respectively. Signals were detected using the SuperSignalWest Pico Chemiluminescent Substrate (Pierce, Rockford, IL) and Amersham ECL hyperfilm (GE Healthcare).
Results
Selection Criteria for Reconstruction of Extinct L1 Elements
Two criteria were considered before selecting the particular L1 subfamily members to reconstruct. First, we focused on the period of high Alu activity when competition with L1 may have been intense. Amplification of the Alu J and S subfamilies (fig. 1) contributed approximately 850,000 copies, accounting for the majority (∼80%) of the Alu elements currently present in the human genome (Shen et al. 1991). The dominant L1 subfamilies that existed during the different periods of individual Alu subfamily activity range from L1PA13 to L1PA1 (fig. 1), with L1PA8 being active during the peak of Alu insertion and the expansion of the Alu S subfamilies approximately 40 Ma (Ohshima et al. 2003). Therefore, we selected L1PA8 as one of two ancestral elements for reconstruction.
Our second criterion was based on observations of rapid protein sequence evolution during L1 subfamily progression. A previous L1 subfamily study (Khan et al. 2006) evaluated the ratio between the fixation rates of nonsynonymous (Ka) and synonymous (Ks) mutations on the derived consensus sequences of the different L1 subfamilies and determined that the coding sequences of ORF2 have remained relatively conserved across subfamilies. However, they show that ORF1 experienced a long spell of positive selection ranging from ∼12 to 40 Ma, with particularly high protein evolution approximately 15–20 Ma during the transition from L1PA5 as the dominant subfamily to L1PA3. We re-evaluated these data using the published consensus sequences (Khan et al. 2006) but updated with changes to the L1PA8 consensus described in the next section, by implementing a similar Ka/Ksanalysis (table 1) on ORF1 and ORF2, with particular focus on the different regions of the L1 ORF2 protein. Unlike the previous study, we subdivided the ORF2p into four distinct regions: the endonuclease domain (endo), the region between the endonuclease and RT (inter endo-RT), RT domain, and the carboxy terminus containing the “zinc-knuckle” cysteine-rich domain (cys). Our analysis confirmed the previous observations that ORF2p generally shows signs of purifying selection when using the full-length sequence for the analysis. However, in contrast to other regions and changes between other L1 subfamilies, the cys domain experienced a notable increase in amino acid substitution rate at approximately 18–20 Ma, during the transition from the L1PA5 to the L1PA4 subfamilies (table 1, bold font). Interestingly, this rapid protein evolution appears to have occurred during an evolutionary time frame that coincides with the ending of a long period of highly permissive L1 trans-mobilization of Alu and processed pseudogenes and the emergence of the SVA retroelement (Ohshima et al. 2003; Wang et al. 2005). The concurrence of ORF2p cys domain evolution with changes in ORF1p is noteworthy; but any relationship between the two observations can only be speculative at this time. Thus, on the basis of these data, we decided to reconstruct L1PA4 as our second selection. Together with L1PA8, we have two ancestral L1 elements that contain the ancestral (L1PA8) and derived (L1PA4) protein sequences spanning this notable period of rapid evolution and that coincided with the observed changes in Alu subfamily evolution (fig. 1).
Table 1.
Analysis of Nonsynonymous (Ka) versus Synonymous (Ks) Substitutions of the Consensus Sequence of the Individual ORF2 Domains of the L1PA Family.
| L1 Pair |
Ka/Ks Analyses on L1
ORF Domains (Nucleotide Positions from Consensus Sequences)a |
|||||
|---|---|---|---|---|---|---|
| ORF1 Full Length (31–1,053) | ORF2 Full Length (1,120–4,944) | ORF2 Endo Domain (1,120–1,836) | ORF2 Inter Endo-RT Domain (1,837–2,649) | ORF2 RT Domain (2,650–3,438) | ORF2 Cys Domain (3,439–4,944) | |
| L1PA2 → L1PA1 | 0.2749 | 0.1951 | 0.2927 | 0.1638 | 0.0558 | 0.2371 |
| L1PA3 → L1PA2 | 0.2738 | 0.2473 | 0.8786 | 0.1806 | 0.1861 | 0.1774 |
| L1PA4 → L1PA3 | 1.4588 | 0.0846 | 0.0486 | 0.0675 | 0.0000 | 0.1906 |
| L1PA5 → L1PA4 | 2.4666 | 0.2551 | 0.0728 | 0.0000 | 0.0000 | 1.5992 |
| L1PA6 → L1PA15 | 0.3911 | 0.1443 | 0.2429 | 0.1201 | 0.0866 | 0.1775 |
| L1PA7 → L1PA6 | 0.6115 | 0.1914 | 0.3219 | 0.2740 | 0.0869 | 0.1635 |
| L1PA8 → L1PA7 | 0.4187 | 0.2245 | 0.2873 | 0.5556 | 0.2829 | 0.0884 |
| L1PA8A → L1PA8 | 0.2256 | 0.2068 | 0.3998 | 0.1974 | 0.1003 | 0.2125 |
| L1PA10 → L1PA8A | 0.3260 | 0.1272 | 0.2903 | 0.1439 | 0.0128 | 0.1249 |
| L1PA11 → L1PA10 | 0.4537 | 0.1708 | 0.0862 | 0.3012 | 0.0518 | 0.2127 |
| L1PA13B → L1PA11 | 0.3500 | 0.1936 | 0.4137 | 0.1849 | 0.0337 | 0.2428 |
| L1PA12 → L1PA13B | 0.4300 | 0.1813 | 0.2669 | 0.2266 | 0.0359 | 0.2170 |
| L1PA13 → L1PA12 | 0.2204 | 0.2530 | 0.3845 | 0.2688 | 0.0642 | 0.2994 |
Note.—Numbers in bold indicate an increase in amino acid substitution rate. aConsensus sequences from Khan et al. (2006).
Construction of Extinct L1 Elements
We used the published L1PA4 and L1PA8 consensus (Khan et al. 2006) to generate the presumed ancestral sequences for our reconstructed L1 elements. This method is not ideal for typical gene trees where older substitutions tend to outnumber younger substitutions in samples of extant sequences. However, given the unique evolutionary dynamics of retroelements, L1 gene trees resemble star phylogenies with a few active elements within a subfamily giving rise to numerous additional copies (Arndt et al. 2003). Thus, sampling biases generated by substitution timeframes should have a negligible effect on the assumption that ancestral L1 subfamily sequences are likely to resemble the consensus sequence of human reference assembly genomic copies corrected for CpG mutations. Another concern is that most of the L1 sequence data available for alignments consists of 5′-truncated elements, making it more difficult to generate reliable consensus sequence for ORF1p and the N-terminus of the ORF2p. Thus, particular attention was given to these regions during the verification of the consensus sequences.
L1 elements generate limited amounts of full-length RNA due to internal splice sites (Belancio et al. 2008), internal pAs (Perepelitsa-Belancio and Deininger 2003), and overall A-richness (Han et al. 2004), making it difficult to quantitatively differentiate L1 subfamilies with respect to cis-retrotransposition rates and trans-mobilization of Alu. These factors could potentially also lead to the translation of differing amounts of ORF1p and ORF2p. We wished to specifically characterize any role(s) that protein sequence differences might have between subfamilies. Therefore, we codon optimized consensus sequences to reconstruct extinct L1 elements with unchanged amino acid sequences but with strategic changes at synonymous codon positions to reduce transcriptional and translational variation between elements. Codon-optimized L1 elements have previously been created with amino acid sequences identical to active modern human and rodent elements (Han and Boeke 2004; Wagstaff et al. 2011). In these published cases, the synthetic L1s appear to be comparable to wild-type L1s in a cultured cell retrotransposition assay but have higher retrotransposition efficiencies when compared with the equivalent wild-type L1 elements. For the design and synthesis of our synthetic L1PA4 and L1PA8 full-length and ORF2 alone constructs, we followed the same codon optimization and plasmid assembly strategy we previously used for the synthesis of L1PA1 (see Materials and Methods) (Wagstaff et al. 2011). To add further evidence of functionality, we also reconstructed a wild-type (nonoptimized) L1PA8 construct for analysis and comparison alongside the synthetic version.
The synthetic L1PA8 and L1PA4 consensus sequences were cloned into constructs that would either support expression of the ORF2 protein, the expression of the full-length L1 (untagged), or the expression of an L1 with a neomycin cassette (tagged) that would allow evaluation of retrotransposition in a culture assay system (fig. 2). We initially tested the retrotransposition competence of the ORF2 constructs by assaying their ability to support Alu retrotransposition in cultured HeLa cells and found that the consensus L1PA8 ORF2 was unable to drive Alu retrotransposition. Given that Alu retrotransposition is readily supported by both human and rodent L1 ORF2 sources, including chimeric human–rodent ORF2s (Wagstaff et al. 2011), we decided to re-evaluate the L1PA8 consensus sequence. Because current L1PA8 human genome copies are often truncated and highly battered, we sought to determine whether manual editing of the sequence could correct errors that emerge from automated consensus building.
To identify genomic copies of L1PA8, we used the published L1PA8 consensus as a BLAT query (UCSC Genome Browser, hg19 Assembly: http://genome.ucsc.edu/cgi-bin/hgBlat) and identified 23 genomic copies that were full-length or near-full-length elements annotated as L1PA8. To validate these L1 copies as belonging to the L1PA8 subfamily, we queried these 23 copies to Repeat Masker (http://www.repeatmasker.org/). Repeat Masker identified 13 copies as L1PA8 and 10 of the copies as belonging to subfamilies other than L1PA8. Thus, for our final L1PA8 set, we only used the 13 L1s that Repeat Masker confirmed as L1PA8 for our subsequent consensus analysis.
The alignment of these 13 L1PA8 sequences led to a modified consensus sequence with 11 amino acid changes in the ORF2 sequence relative to the original consensus. These 11 codon positions are shown for each of the 13 L1PA8 sequences in table 2. In all cases, our modified consensus is supported by a plurality of the individual sequences. Table 3 lists the individual changes made to the modified consensus and the rationale for those changes. Because the CpG dinucleotides mutate at a rate that is approximately 10 times faster than non-CpG positions as a result of the deamination of 5-methylcytosine (Bird 1980), we specifically searched for changes associated with CpGs. Four of the 11 codons contain CpG correction errors, and the remaining codons were either polymorphic or supported by each of the individual sequences from our alignment. An example of an ambiguous amino acid in the ORF2p from L1PA8 is shown in figure 3. There are several possible explanations for the differences between our modified consensus and the original published L1PA8 ORF2 consensus sequence: 1) we used different individual elements to construct the consensus sequence, 2) uncertain alignments, particularly with respect to small deletions and adjacent nucleotides, and 3) ascertaining CpG sites. We had the additional benefit of closely scrutinizing the differences between the modified and original consensus sequences. Comparison to the closest ancestral (L1PA8A) and derived (L1PA7) subfamilies of L1PA8 provides further support for the 11 codon modifications we made (last two rows of table 2). Before the corrections, 10 of the 11 codons were not shared by either the ancestral or derived subfamilies. Following the modifications, 9 of the 11 codons match the corresponding codon for both of these subfamilies, whereas the remaining two codons match one related subfamily. Therefore, these changes are the most parsimonious with respect to sequence polymorphisms and evolutionary progression of subfamilies. A complete sequence alignment of the amino acids changed for ORF2 PA8 is shown in supplementary figure S1, Supplementary Material online. We used similar precautionary measures but identified no amino acids to modify for the ORF1 PA8 nor the ORF1 and ORF2 of the L1PA4 consensus sequence. Our wild-type L1PA8 construct also contains these 11 modified amino acids. We assembled the L1 and Alu sequences into tagged and/or untagged constructs (fig. 2) to evaluate cis- and trans-mobilization in cultured HeLa cells.
Table 3.
Rationale for Changes to the L1PA8 Consensus Sequence.
| Positiona | Consensus AA | Modified AA | Support for Choice of Modification |
|---|---|---|---|
| 47 | M | T | Most common residue + CpG site |
| 101 | N | K | Most common residue + polymorphic site |
| 104 | T | M | Most common residue + CpG site |
| 347 | V | L | Most common residue + polymorphic site |
| 375 | G | R | Most common residue + polymorphic site |
| 716 | P | Q | Most common residue + polymorphic site |
| 755 | N | S | Most common residue + polymorphic site |
| 777 | M | V | Most common residue + polymorphic site |
| 838 | A | T | Most common residue + CpG site |
| 918 | N | D | Most common residue + CpG site |
| 1092 | V | M | Most common residue + polymorphic site |
aAmino acid sequence numbers using the ORF2 L1RP sequence as reference.
Fig. 3.
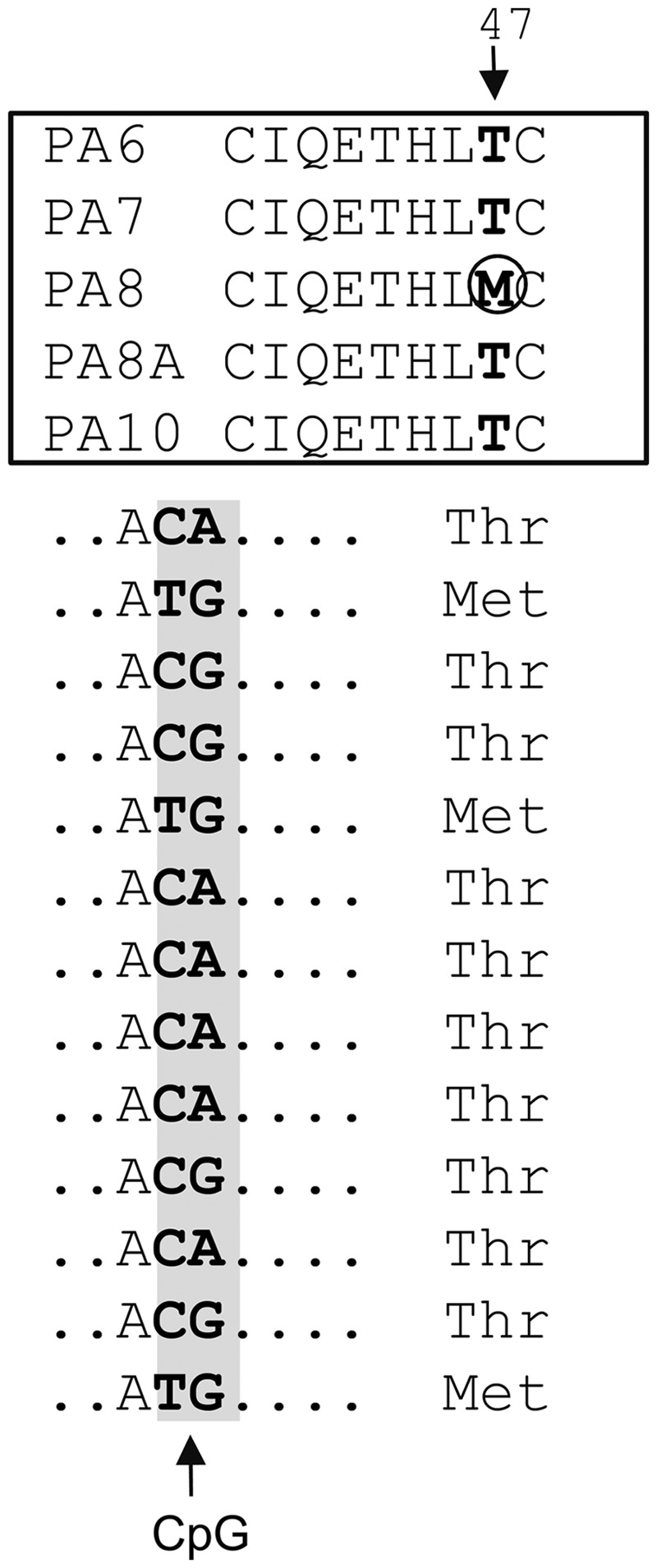
Revision of the L1PA8 sequence. Example of the approach used in the identification of L1PA8 consensus codon sequences conforming to the criteria for modification. The top panel shows an alignment of the amino acid sequences positions 40–48 of the published consensus sequences L1PA subfamilies (Khan et al. 2006) that allowed for the identification of methionine (M) at codon 47 of the ORF2 protein (circled) to be a potential L1PA8-specific change. The bottom panel shows a nucleotide sequence alignment of ORF2 protein codon 47 (flanking sequence is represented by dots) from our subset of full-length L1PA8 copies. The sequences are highly variable due to the presence of a CpG (C to T or G to A). The original L1PA8 consensus had a methionine at this position due to a CpG correction error. However, the alignment of our 13 L1PA8 copies supports the threonine codon as the most likely to have been present in the active L1PA8 element. The presence of a threonine is further supported by the observation that the other L1PA subfamilies in the time periods flanking L1PA8 also contain a threonine (T) at this position. Using these criteria, we corrected codon 47 of ORF2 of the L1PA8 consensus.
Evaluation of the Reconstructed L1s
The reconstructed full-length L1PA4 and L1PA8 elements proved to be retrocompetent in HeLa cells (fig. 4A). Our optimized version of the L1PA1 element has previously been shown to be highly retrocompetent and more active than wild-type L1 in cultured cells (Wagstaff et al. 2011). The optimized L1PA8 element shows a slightly higher retrotransposition efficiency relative to L1PA1 (∼125%, paired t-test P < 0.001). As with previous comparisons between optimized and wild-type L1 elements, the optimized version of L1PA8 is more active in this assay than its wild-type counterpart (paired t-test P < 0.001). Considering that the L1PA1 is the optimized version of the most active human L1 reported, the L1RP, this indicates that both our optimized L1PA4 and L1PA8 constructs are highly efficient.
We performed two separate controls to confirm that the colonies from the L1PA4 and L1PA8 transfections represented genuine retrotransposition events. First, we harvested HeLa DNA from colony pools and showed by PCR analysis that L1PA4 and L1PA8 inserts contain the resistance tag with the intron spliced out (fig. 4B, top panel). Because splicing only occurs in transcripts generated by the CMV promoter of our tagged L1 constructs, this confirms that the antibiotic resistance is not due to protein expression from unincorporated plasmid in transfected cells. We further show that colony formation does not occur in the presence of the RT inhibitor, d4t (fig. 4B, bottom panel), which has previously been shown to effectively inhibit L1 retrotransposition in HeLa cells (Kroutter et al. 2009).
The codon-optimized neomycin L1-tagged constructs generated equivalent amounts of spliced full-length L1 transcripts (fig. 4C). As expected, the wild-type constructs (PA8wt and L1.3 wt) have lower transcription levels than the optimized versions. Although there is approximately a 30-fold difference in the amount of transcript generated between the codon-optimized and the wild-type constructs, retrotransposition rates only differ by ∼7.4 fold for L1PA8 and ∼2.2 fold for L1PA1, indicating a nonlinear relationship between the amount of L1 RNA and insertional capability, as has previously been observed (An et al. 2011).
Transmobilization of Old and Young Alu Subfamilies
We generated a set of tagged Alu constructs comprising the consensus sequences of the young currently active subfamilies (Alu Ya5 and Alu Y), an intermediate (Alu Sg1, previously known as Alu “AS” [Shen et al. 1991; Batzer et al. 1996]), and two older subfamilies (Alu Sx and Alu Jo). Expression analysis of the Alu constructs demonstrates equivalent expression between all the tagged Alu subfamily transcripts (fig. 5A). We also verified that the RNA and protein (ORF1p) expression levels of the driver L1s were equivalent for the vectors of the three different L1 subfamilies (fig. 5B). We next evaluated these modern and ancestral retroelement constructs to test for variation in Alu retrotransposition efficiency when driven by the different L1 subfamilies in culture. Because Alu only requires ORF2p for retrotransposition (Dewannieux et al. 2003; Wallace et al. 2008), we first chose to evaluate the effect of L1PA1, L1PA4, and L1PA8 ORF2p on Alu subfamily activity (fig. 5C). Under these conditions, our negative controls showed no background (G418 resistant colonies) when the Alu construct was not supplemented with ORF2p (supplementary figs. S3 and S4, Supplementary Material online). The younger Alu elements consistently showed higher retrotransposition efficiency than the older Alu Jo when driven by the ORF2p of the younger L1s (PA1 and PA4; P < 0.001). However, there are no significant differences in Alu subfamily activity when the ORF2p of L1PA8 drives retrotransposition. Instead, retrotransposition efficiency of the younger Alu elements decreases to levels comparable to Alu Jo (supplementary fig. S3A, Supplementary Material online). These results are consistently observed even when varying transfection conditions by using different Alu/ORF2 ratios (supplementary fig. S4, Supplementary Material online). Performing the Alu subfamily retrotransposition analysis using full-length optimized L1 elements to drive retrotransposition showed similar results (fig. 5D) but with a lower retrotransposition efficiency (supplementary fig. S3B, Supplementary Material online). Under these conditions, the difference in retrotransposition efficiency between Alu Jo and the younger Alu subfamilies was only observed with L1PA1. Although the Alu Sg1 (∼25–35 Ma) shows a trend for a higher retrotransposition rate relative to the other Alu subfamilies, due to the intrinsic experimental variability, it is not significantly different (P = 0.385).
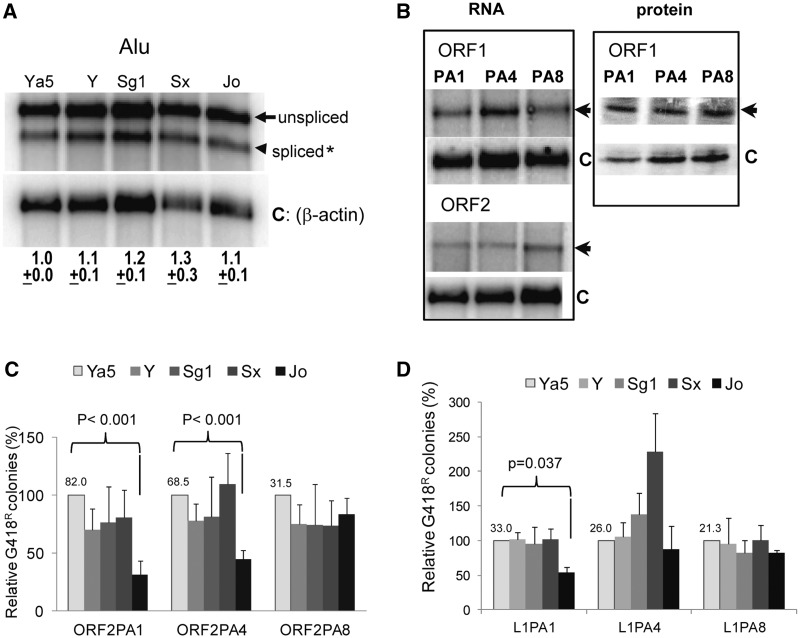
Fig. 5.
L1 PA4 and L1 PA8 support retrotransposition of ancestral Alu subfamilies. (A) Evaluation of the RNA profiles of the different tagged Alu subfamily constructs. Northern blot analysis of poly-A selected RNA extracts was performed from HeLa cells transiently transfected with the tagged constructs of five different Alu subfamilies that were active during distinct evolutionary periods. The unspliced (arrow) and spliced (arrowhead) neo-tagged Alu transcripts are indicated. The spliced Alu transcripts were normalized to β-actin (C, loading control) and expressed relative to the AluYa5 that was arbitrarily designated as 1.0. The mean ± standard error of the mean (SEM) for the quantitation results for each construct are indicated below (n = 3). No significant differences were observed between the Alu subfamily transcripts (one-way analysis of variance, F = 0.46, P = 0.763722; n = 3). (B) Evaluation of RNA expression from the ORF1 or ORF2 constructs. Poly-A selected RNA and protein levels were evaluated from HeLa cells transiently transfected with the codon-optimized ORF1 and ORF2 protein expression vectors from the different L1 subfamilies (PA1, PA4, and PA8). RNA blots were hybridized with a riboprobe complementary to the 3′-region of the ORF1/ORF2 transcript indicated by the arrow and a riboprobe complementary to β-actin mRNA as control (C). Extracts were obtained from HeLa cells transiently transfected with the codon-optimized myc-tagged L1 ORF1 protein expression vector from the different subfamilies (PA1, PA4, and PA8). Protein blots were incubated with anti-myc indicated by the arrow and anti-β actin as control (C). (C) Retrotransposition of the tagged consensus Alu subfamilies (Ya5, Yb9, Sg1, Sx, and Jo) driven by the ORF2 protein of the different L1 subfamilies (L1PA1, L1PA4, and L1PA8). The Alu Ya5 data were used to define 100%. The mean number of G418R before normalization for the AluYa5 is shown above the column. Columns and error bars represent the % mean G418R colonies ± SEM. P values indicate that the retrotransposition efficiency of the older Alu element (AluJo) is significantly lower than the modern AluYa5 when the ORF2 driver is from L1PA1 or L1PA8 (paired t-test, P < 0.001). (D) Retrotransposition of the tagged consensus Alu subfamilies (Ya5, Y, Sg1, Sx, and Jo) driven by the full-length L1 of the different subfamilies (L1PA1, L1PA4, and L1PA8). The mean number of G418R before normalization for the AluYa5 is shown above the column. Columns and error bars formatted as in C. P values indicate that the retrotransposition efficiency of the older Alu element (AluJo) is significantly lower than the modern AluYa5 when L1PA1 is the driver element (paired t-test, P = 0.037; n ≥ 3).
Discussion
Our data demonstrate that the use of consensus L1 sequences is a viable approach for the reconstruction of extinct L1 subfamilies. However, our initial failure to produce a retrocompetent L1PA8 ORF2 sequence demonstrated the limitations to the approach, particularly for older subfamilies. The primary stumbling block is the reliability of the data used to derive the consensus sequence. In particular, the nucleotide changes caused by the deamination of methylated CpGs present in the sequences used to build the consensus require careful attention. In the case of L1PA8 ORF2, 4 out of the 11 identified amino acid changes could be attributed to CpG derived sequence changes. The linear progression of L1 subfamilies provides an additional layer for the analysis of L1 consensus sequences. By comparing temporally adjacent subfamilies (i.e., closely related), amino acid substitutions that appear as singletons (not present in ancestral or derived subfamilies) can be closely scrutinized to make sure CpG or polymorphism correction errors do not occur.
The insertional history of L1 and Alu in primate genomes consists of a linear progression of subfamilies, with only brief temporal overlaps between ancestral subfamilies and the derived subfamilies that replace them. Previous phylogenetic and genetic distance analyses of ancestral LINEs and SINEs (Shen et al. 1991; Ohshima et al. 2003; Khan et al. 2006; Bennett et al. 2008) have shown that insertion rates vary over time, with some subfamilies reaching much higher copy numbers than others. There is no indication of a positive correlation for insertion rate between LINEs and SINEs across evolutionary time, suggesting that if there were lenient and restrictive insertional time periods, those periods were not the same for L1 and Alu. Instead, the historical amplification patterns of L1 and Alu suggest a possible negative relationship, with L1 showing a relatively high insertion rate that only decreases with the emergence and proliferation of Alu (fig. 1). Peak Alu amplification also coincides with peak formation of processed pseudogenes (Ohshima et al. 2003). This may indicate a period of general genomic leniency for new genomic inserts, except that the corresponding L1 insertion rate is comparatively low. Alternatively, one or more of the active L1 subfamilies from this period may have been especially vulnerable to nonautonomous elements. The period corresponding to the more recent expansion of Alu Y is interesting for a couple of reasons. Peak Alu Y amplification (fig. 1) corresponds with the emergence and proliferation of the nonautonomous SVA retroelement ∼18–25 Ma (Wang et al. 2005) and the rapid evolution of ORF1p and ORF2p during the transition from L1PA5 to L1PA4 (table 1). Whether both L1 proteins evolved in response to Alu and/or SVA competition, host factors, or other evolutionary pressures remains to be determined.
There is a slight indication of differential interaction between younger L1 elements and the different Alu subfamilies. However, the small observed difference between modern and ancestral L1 elements is less likely, on its own, to explain the changing insertional dynamics of Alu amplification. Other explanations to the evolutionary pattern of Alu amplification exist. The Alu “master” or “source” element model suggests the existence of a small number of hyperactive source elements that are responsible for the accumulation of the new Alu copies (Deininger et al. 1992). Stochastic changes in the number of source elements during any given time period could be a factor in determining Alu amplification patterns. In addition, Alu amplification dynamics may have been significantly influenced by “stealth-driver” elements (Han et al. 2005), with the appearance of short-lived hyperactive copies regulating Alu amplification dynamics. This pattern is apparent in the analysis of the Orangutan genome. The low number of Orangutan-specific Alu insertions may be because of low “stealth” Alu amplification (Walker et al. 2012) in a genome lacking short-lived hyperactive Alu copies. Thus, the combination of population dynamics and stochastic variation in active Alu elements has likely played a role in Alu subfamily proliferation and evolution.
A limitation to the investigation of ancestral LINE and SINE elements is the inability to replicate the exact cellular environments that existed during their proliferation. Any interactions between LINEs and SINEs are likely to be mediated by cellular factors and those interactions could well be lost in living tissues and immortalized cell lines. Multiple studies show that endogenous retroelement activity can be regulated by cellular factors (reviewed in Levin and Moran 2011). Examples include, the human APOBEC3 family of cytidine deaminases (Bogerd et al. 2006), the MOV10 superfamily 1 putative RNA helicase (Arjan-Odedra et al. 2012), the 3′-repair exonuclease 1, TREX1 (Stetson et al. 2008), and “flap” endonuclease XPF/ERCC1 heterodimer (Gasior et al. 2008). In addition, different interfering RNA-based mechanisms, including siRNAs and piRNAs, have been shown to inhibit mobile elements (reviewed in Levin and Moran 2011). Because of the possibility for coevolution with parasitic mobile elements, host factors may evolve rapidly, leading to changing cellular environments. For example, antagonistic interactions between primates and their retroviruses or retroelements can lead to rapid evolution of host factors to limit their proliferation. Several recent studies have shown that APOBEC genes have evolved rapidly in human ancestors and differentially regulate retrovirus and/or retroelement activity in primates (OhAinle et al. 2006; Stenglein and Harris 2006; Niewiadomska et al. 2007; Tan et al. 2009; Duggal et al. 2011). These interactions can lead to a state of perpetual coevolution between cellular factors and pathogens. Whether APOBEC genes directly target retroelements or affect them indirectly because of their interaction with retroviruses is undetermined. Although Alu requires L1 proteins to retrotranspose, there are examples of some factors that differentially affect L1 and SINE mobilization (Hulme et al. 2007; Kroutter et al. 2009; Ichiyanagi et al. 2011). Our inability to measure any major differential interactions between ancestral LINE and SINE subfamilies could simply be because the mediating cellular factors are no longer active in modern humans.
Either way, the historical activity of LINEs and SINEs has likely been influenced by host factors that evolve to combat changing cellular threats and stochastic events that affect the number of active elements at any given period. We are currently evaluating the influence of cellular factors on LINE and/or SINE subfamily activity.
Supplementary Material
Supplementary figures S1–S4 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
Full-length L1 consensus sequences were kindly provided by Stéphane Boissinot. The authors thank Beibei Xu for helping to assemble the pBS-L1PA8WTmneo construct. This work was supported by the National Institutes of Health R01 GM079709-01 to A.M.R-E.; P20 P20GM103518/PRR020152 to A.M.R-E. and V.P.B.; 5K01AG030074-02 to V.P.B.; and the Ellison Medical Foundation New Scholar in Aging award [547305G1 to V.P.B.].
References
- Alemán C, Roy-Engel AM, Shaikh TH, Deininger PL. Cis-acting influences on Alu RNA levels. Nucleic Acids Res. 2000;28:4755–4761. doi: 10.1093/nar/28.23.4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An W, Dai L, Niewiadomska AM, Yetil A, O'Donnell KA, Han JS, Boeke JD. Characterization of a synthetic human LINE-1 retrotransposon ORFeus-Hs. Mob DNA. 2011;2:2e. doi: 10.1186/1759-8753-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arjan-Odedra S, Swanson CM, Sherer NM, Wolinsky SM, Malim MH. Endogenous MOV10 inhibits the retrotransposition of endogenous retroelements but not the replication of exogenous retroviruses. Retrovirology. 2012;9:53. doi: 10.1186/1742-4690-9-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt PF, Petrov DA, Hwa T. Distinct changes of genomic biases in nucleotide substitution at the time of mammalian radiation. Mol Biol Evol. 2003;20:1887–1896. doi: 10.1093/molbev/msg204. [DOI] [PubMed] [Google Scholar]
- Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat Rev Genet. 2002;3:370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- Batzer MA, Deininger PL, Hellmann-Blumberg U, Jurka J, Labuda D, Rubin CM, Schmid CW, Zietkiewicz E, Zuckerkandl E. Standardized nomenclature for Alu repeats. J Mol Evol. 1996;42:3–6. doi: 10.1007/BF00163204. [DOI] [PubMed] [Google Scholar]
- Belancio VP, Hedges DJ, Deininger P. LINE-1 RNA splicing and influences on mammalian gene expression. Nucleic Acids Res. 2006;34:1512–1521. doi: 10.1093/nar/gkl027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belancio VP, Roy-Engel AM, Deininger P. The impact of multiple splice sites in human L1 elements. Gene. 2008;411:38–45. doi: 10.1016/j.gene.2007.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett EA, Keller H, Mills RE, Schmidt S, Moran JV, Weichenrieder O, Devine SE. Active Alu retrotransposons in the human genome. Genome Res. 2008;18:1875–1883. doi: 10.1101/gr.081737.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird AP. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 1980;8:1499–1504. doi: 10.1093/nar/8.7.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd H, Wiegand HL, Hulme AE, Garcia-Perez JL, O'Shea KS, Moran JV, Cullen BR. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc Natl Acad Sci U S A. 2006;103:8780–8785. doi: 10.1073/pnas.0603313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissinot S, Furano AV. Adaptive evolution in LINE-1 retrotransposons. Mol Biol Evol. 2001;18:2186–2194. doi: 10.1093/oxfordjournals.molbev.a003765. [DOI] [PubMed] [Google Scholar]
- Cost GJ, Feng Q, Jacquier A, Boeke JD. Human L1 element target-primed reverse transcription in vitro. EMBO J. 2002;21:5899–5910. doi: 10.1093/emboj/cdf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning AP, Gu W, Castoe TA, Batzer MA, Pollock DD. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011;7:e1002384. doi: 10.1371/journal.pgen.1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger PL, Batzer MA, Hutchinson ICA, Edgell MH. Master genes in mammalian repetitive DNA amplification. Trends Genet. 1992;8:307–311. doi: 10.1016/0168-9525(92)90262-3. [DOI] [PubMed] [Google Scholar]
- Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- Dombroski BA, Scott AF, Kazazian HH., Jr Two additional potential retrotransposons isolated from a human L1 subfamily that contains an active retrotransposable element. Proc Natl Acad Sci U S A. 1993;90:6513–6517. doi: 10.1073/pnas.90.14.6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggal NK, Malik HS, Emerman M. The breadth of antiviral activity of apobec3DE in chimpanzees has been driven by positive selection. J Virol. 2011;85:11361–11371. doi: 10.1128/JVI.05046-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault C, Casella JF, Heidmann T. A Tetrahymena thermophila rybozyme-based indicator gene to detect transposition of marked retroelements in mammalian cells. Nucleic Acids Res. 2002;30:e49. doi: 10.1093/nar/30.11.e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Moran JV, Kazazian HH, Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- Gasior SL, Roy-Engel AM, Deininger PL. ERCC1/XPF limits L1 retrotransposition. DNA Repair. 2008;7:983–989. doi: 10.1016/j.dnarep.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JS, Boeke JD. A highly active synthetic mammalian retrotransposon. Nature. 2004;429:314–318. doi: 10.1038/nature02535. [DOI] [PubMed] [Google Scholar]
- Han JS, Szak ST, Boeke JD. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature. 2004;429:268–274. doi: 10.1038/nature02536. [DOI] [PubMed] [Google Scholar]
- Han K, Xing J, Wang H, Hedge DJ, Garber RK, Cordaux R, Batzer MA. Under the genomic radar: the stealth model of Alu amplification. Genome Res. 2005;15:655–664. doi: 10.1101/gr.3492605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges DJ, Callinan PA, Cordaux R, Xing J, Barnes E, Batzer MA. Differential alu mobilization and polymorphism among the human and chimpanzee lineages. Genome Res. 2004;14:1068–1075. doi: 10.1101/gr.2530404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulme AE, Bogerd HP, Cullen BR, Moran JV. Selective inhibition of Alu retrotransposition by APOBEC3G. Gene. 2007;390:199–205. doi: 10.1016/j.gene.2006.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichiyanagi K, Li Y, Watanabe T, et al. (13 co-authors) Locus- and domain-dependent control of DNA methylation at mouse B1 retrotransposons during male germ cell development. Genome Res. 2011;21:2058–2066. doi: 10.1101/gr.123679.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan H, Smit A, Boissinot S. Molecular evolution and tempo of amplification of human LINE-1 retrotransposons since the origin of primates. Genome Res. 2006;16:78–87. doi: 10.1101/gr.4001406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroutter EN, Belancio VP, Wagstaff BJ, Roy-Engel AM. The RNA polymerase dictates ORF1 requirement and timing of LINE and SINE retrotransposition. PLoS Genet. 2009;5:e1000458. doi: 10.1371/journal.pgen.1000458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, et al. (256 co-authors) Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Levin HL, Moran JV. Dynamic interactions between transposable elments and their hosts. Nat Rev Genet. 2011;12:615–627. doi: 10.1038/nrg3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Cherukuri PF, et al. (24 co-authors) CDD: a conserved domain database for protein classification. Nucleic Acids Res. 2005;33:D192–D196. doi: 10.1093/nar/gki069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewiadomska AM, Tian C, Tan L, Wang T, Sarkis PTN, Yu XF. Differential inhibition of long interspersed element 1 by APOBEC3 does not correlate with high-molecular-mass-complex formation or P-body association. J Virol. 2007;81:9577–9583. doi: 10.1128/JVI.02800-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OhAinle M, Kerns JA, Malik HS, Emerman M. Adaptive evolution and antiviral activity of the conserved mammalian cytidine deaminase APOBEC3H. J Virol. 2006;80:3853–3862. doi: 10.1128/JVI.80.8.3853-3862.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima K, Hattori M, Yada T, Gojobori T, Sakaki Y, Okada N. Whole-genome screening indicates a possible burst of formation of processed pseudogenes and Alu repeats by particular L1 subfamilies in ancestral primates. Genome Biol. 2003;4:R74. doi: 10.1186/gb-2003-4-11-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perepelitsa-Belancio V, Deininger PL. RNA truncation by premature polyadenylation attenuates human mobile element activity. Nat Genet. 2003;35:363–366. doi: 10.1038/ng1269. [DOI] [PubMed] [Google Scholar]
- Rinehart TA, Grahn RA, Wichman HA. SINE extinction preceded LINE extinction in sigmodontine rodents: implications for retrotranspositional dynamics and mechanisms. Cytogenet Genome Res. 2005;110:416–425. doi: 10.1159/000084974. [DOI] [PubMed] [Google Scholar]
- Sassaman DM, Dombroski BA, Moran JV, Kimberland ML, Naas TP, DeBerardinis RJ, Gabriel A, Swergold GD, Kazazian HH., Jr Many human L1 elements are capable of retrotransposition. Nat Genet. 1997;16:37–43. doi: 10.1038/ng0597-37. [DOI] [PubMed] [Google Scholar]
- Shen MR, Batzer MA, Deininger PL. Evolution of the master Alu gene(s) J Mol Evol. 1991;33:311–320. doi: 10.1007/BF02102862. [DOI] [PubMed] [Google Scholar]
- Smit AF. Interspersed repeats and other mementos of transposable elmenets in mammalian genomes. Curr Opin Genet Dev. 1999;9:657–663. doi: 10.1016/s0959-437x(99)00031-3. [DOI] [PubMed] [Google Scholar]
- Smit AFA, Toth G, Riggs AD, Jurka J. Ancestral, mammalian-wide subfamilies of LINE-1 repetitive sequences. J Mol Biol. 1995;26:401–417. doi: 10.1006/jmbi.1994.0095. [DOI] [PubMed] [Google Scholar]
- Stenglein MD, Harris RS. APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. J Biol Chem. 2006;281:16837–16841. doi: 10.1074/jbc.M602367200. [DOI] [PubMed] [Google Scholar]
- Stetson DB, Ko JS, Heidmann T, Medhiov R. Trex1 prevents cell-intrincsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Sarkis PT, Wang T, Tian C, Yu XF. Sole copy of Z2-type human cytidine deaminase APOBEC3H has inhibitory activity against retrotransposons and HIV-1. FASEB J. 2009;23:279–287. doi: 10.1096/fj.07-088781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagstaff BJ, Barnerssoi M, Roy-Engel AM. Evolutionary conservation of the functional modularity of primate and murine LINE-1 elements. PLoS One. 2011;6:e19672. doi: 10.1371/journal.pone.0019672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JA, Konkel MK, Ullmer B, Monceaux CP, Ryder OA, Hubley R, Smit AFA, Batzer MA. Orangutan Alu quiescence reveals possible source element: support for ancient backseat drivers. Mob DNA. 2012;3:8. doi: 10.1186/1759-8753-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace N, Wagstaff BJ, Deininger PL, Roy-Engel AM. LINE-1 ORF1 protein enhances Alu SINE retrotransposition. Gene. 2008;419:1–6. doi: 10.1016/j.gene.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Xing J, Grover D, Hedges DJ, Han K, Walker JA, Batzer MA. SVA elements: a hominid-specific retroposon family. J Mol Biol. 2005;354:994–1007. doi: 10.1016/j.jmb.2005.09.085. [DOI] [PubMed] [Google Scholar]
- Weichenrieder O, Repanas K, Perrakis A. Crystal structure of the targeting endonuclease of the human LINE-1 retrotransposon. Structure. 2004;12:975–986. doi: 10.1016/j.str.2004.04.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.