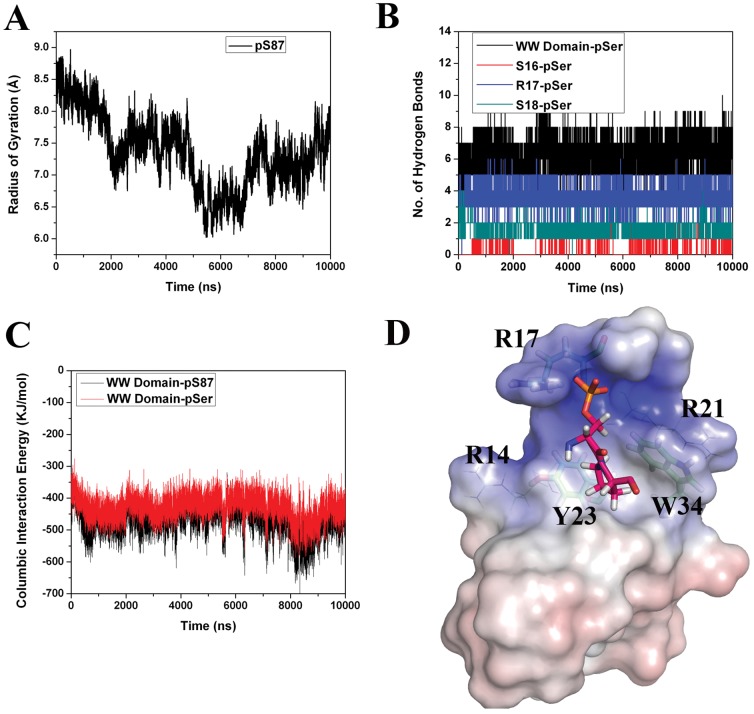
Figure 5. Pin1 WW domain-pS87 MD simulation analyses.
(A) Radius of gyration for pS87 Bcl-2 phosphopeptide in the WW domain-pS87 simulation. (B) Hydrogen bond interaction analysis between pS87 and the WW domain residues. (C) Short range columbic interaction analysis between the WW domain and the pS87, as well as the WW domain and the pSer alone. (D) Electrostatic potential calculations of the WW domain and the pS87 phosphopeptide complex. The arginine residue, which forms a charged/hydrogen bond interaction with pSer, is highlighted as sticks, and the other two arginine residues are represented with lines. The hydrophobic pocket residues (Y23 and W34) are also shown in stick representation. The pSer-Pro motif is also highlighted.