Specific cytokine signatures correlate with genetic aberrations in CLL/SLL, reflecting a pattern of Th1/Th2/Treg dysregulation that may predict aggressive disease, and a need for therapy.
Keywords: B-cell lymphoma, Th1, Th2, sIL-2Rα, genetic profile
Abstract
Early treatment of CLL/SLL does not impact survival-reflecting limitations in detecting progression early and identifying asymptomatic patients likely to benefit from early treatment. Improved understanding of CLL/SLL biology would identify better prognostic/predictive markers. This study attempts to address these issues by determining the relationship between cytokine aberrations and poor clinical outcomes in CLL/SLL in the context of a genetic–based prognostic model. Fifty-nine serum cytokines/chemokines were measured in 28 untreated CLL/SLL patients. Patients were stratified as GR or int/PR using cytogenetics. Comparison of CLL/SLL with 28 HCs revealed increased expression of Th2 cytokines (IL-10, IL-5, sIL-2Rα; P≤0.01) and decreased levels of Th1 cytokines (IL-17, IL-23, IFN-γ; P≤0.003). In a multivariate analysis of GR versus int/PR groups, differential expression of sIL-2Rα maintained significance with increased expression in int/PR CLL/SLL. With median follow-up of 54.3 months after diagnosis, four patients incurred disease progression, with an IL-17/sIL-2Rα model predicting need for treatment in all cases. In summary, specific cytokine signatures are associated with genetically defined aggressive disease and predict need for therapy. This suggests utility in detecting disease progression early, identifying those likely to incur a survival advantage with early treatment, and directing future therapy.
Introduction
CLL/SLL is one of the most common indolent B cell disorders in elderly adults. The pathogenesis of CLL has been attributed to various forms of immune dysregulation: antigenic stimulation with tonic BCR signaling leads to the accumulation of transforming genetic events [1–4]. An altered microenvironment ensues in bone marrow, lymphoid tissue, and/or peripheral blood with the recruitment of specific T cell subsets and stromal cells with shifts in cytokine/chemokine production in all three compartments [5–11]. In CLL, cytokine changes favoring Tregs and Th2 response with suppression of Th1 differentiation have been described [9–11]. This altered microenvironment collectively favors immune evasion and expansion of a dominant malignant clone [1–4, 10]. With accumulation of additional chromosomal aberrations in this albeit slow but proliferating clone, CLL cells may gain autonomy, surviving independently of signals from its microenvironment [1]. The dynamic biology of CLL immunity provides a viable explanation for the heterogeneous clinical course associated with the disease.
There are a number of tools used for prognostication in CLL/SLL, many of which reflect some element of immune dysregulation. For example, aggressive disease can be characterized by an unmuated IgVH, suggesting circumvention of the Th cell or germinal-center maturation process, typical of a normal B cell response to antigenic stimulation [12–14]. Similarly, increased expression of ZAP-70 and/or CD38 derives prognostic significance from their association with augmented BCR-mediated signaling [12, 14, 15]. A cytogenetic-based prognostic model, however, has been most widely adopted as the prognostic tool of choice, largely because of the sensitivity of FISH used to evaluate chromosomal aberrations and the global validation of a this model as a reliable prognostic tool in predicting time to first treatment and disease-specific survival [16–21]. Despite the ability to prognosticate using cytogenetic information, a “watch-and-wait” approach is favored in all asymptomatic patients with early-stage disease, including those with int/PR and PR cytogenetic features. The apprehension to treat such patients early in their disease course stems from failure to alter overall survival outcomes. This reflects the inability to use cytogenetic information to detect progression early, identify patients likely to benefit from therapy with early-stage disease prior to progression, and individualize therapeutic strategies. An improved understanding of the relationship between cytogenetics and CLL immunity may address these short-comings.
Interestingly, the association between cytogenetics and the altered microenvironment has remained largely unexplored, particularly in relation to a comprehensive measure of cytokines/chemokines. Theoretically, alterations in T cell subsets and associated cytokine expression may provide a mechanistic explanation for the aggressive phenotype associated with int/PR and PR cytogenetics. In this study, luminex-based immunoassays were used to measure the expression of 59 cytokines/chemokines in serum samples of patients with CLL/SLL and explored their prognostic significance in relation to aggressive disease defined by genomic risk.
MATERIALS AND METHODS
Subjects and collection of blood samples
Samples were collected from 28 HCs, 25 untreated CLL, and three untreated SLL patients with written consent under an Institutional Review Board-approved protocol. The diagnosis of CLL/SLL was made according to World Health Organization criteria, and all CLL/SLL samples were deemed CD5+ by flow cytometry. Cytogenetics and/or FISH were obtained on the majority of patients (n=24) and used to stratify them as GR (13q) or int/PR (Tri12), normal cytogenetics, 11q, and 17p. Information on ZAP-70 and CD38 status was collected retrospectively, as these prognosticators had not been established at the time of diagnosis and sample collection for some patients (some diagnoses were made as early as 1998).
CLL/SLL patient samples used for this study were collected at least 1 year after diagnosis but prior to treatment initiation. Peripheral blood was collected by venipuncture, and serum was isolated off the clot from these samples and subsequently stored at −80°C. Fifty-nine cytokine/chemokine protein levels were measured in a 96-well format in duplicate with 50 μl serum from each sample using Milliplex, a multiplex Luminex-based technology. To account for expected fluctuations in cytokines, two different serum samples separated in time by 3 or more months were run in 24 of the CLL/SLL patients: 24 patient samples with 13q representing two samples from 12 patients and 23 samples from patients with Tri12, normal cytogenetics, 11q, or 17p, representing one to two samples from 12 patients were used. Cytokine/chemokine assays included IL-1α and -β, -2, -4, -5, -6, -8, -9, -10, -12, -13, -15, -17, and -33, sIL-2Rα), FGF-2, eotaxin, TNF-α, TGF-α, IFN-α2, IFN-γ, TRAIL, G-CSF, GM-CSF, MIP-1δ, MCP-1, IP-10, and VEGF. β2M levels, a reliable prognostic parameter for poor survival in patients with CLL, was also measured. Results were generated using STarStation Software Version 2.3.
Statistical analysis
Cytokine expression was compared among HCs, GR patients, and patients with int/PR cytogenetics, as determined by karyotype analysis and/or FISH. All data were assumed to be nonparametric, and a P ≤ 0.05 was considered significant. With the use of SPSS Version 18, Mann-Whitney tests (for comparison of two groups) and Kruskal-Wallis tests (for comparison of three or more groups) were performed to analyze differences in continuous variables. Bonferroni regression was used for multivariate analyses. All multivariate analyses were performed in the CLL/SLL population alone, as no demographic data were available for HCs. Filter on Volcano Plot analysis was used to compute log FC of each cytokine, and a FC ≥1.5 was chosen as the threshold of significance, with a P ≤ 0.05. For this analysis, Genespring 11.5.1 software was used to convert data into base-2 logarithmic values with median baseline transformation applied across all samples using the formula: FC = 2mean[log2(CLL cytokine)]/2mean[log2(HC cytokine)].
RESULTS
CLL and SLL patients were included in this study (median age of 56 years ranging from 30 to 77; 17 males). All patients with SLL had Stage IV disease with bone marrow involvement at diagnosis. Risk-relevant information, including karyotype and/or FISH, RAI stage, CD38, and ZAP-70 status, was recorded when available and is shown in Table 1. IgVH data were not available on any patients. Median follow-up was 54.3 months after diagnosis. By this time, four patients had received cytotoxic therapy for disease progression, and one patient received rituximab for a comorbid autoimmune condition.
Table 1. Clinical Characteristics of CLL/SLL Patients at Sample Collection.
| Characteristic | Patients with cytokine data (n=28) |
|
|---|---|---|
| No. | % | |
| Age (years) | ||
| Median | 56 (range 30–77) | |
| >60 | 9 | 32 |
| Sex | ||
| Male | 17 | 61 |
| Female | 11 | 39 |
| RAI stage | ||
| 0 | 19 | 68 |
| 1 | 5 | 18 |
| 2 | 1 | 3 |
| 3 | 0 | 0 |
| 4 | 0 | 0 |
| N/A | 3 (SLL patients) | 11 |
| Absolute lymphocyte count, ×109/l | ||
| <5000 | 3 (SLL patients) | 11 |
| 5000–10,000 | 9 | 32 |
| >10,000 | 16 | 57 |
| Hemoglobin, g/dl | ||
| <11 | 1 | 3 |
| >11 | 27 | 97 |
| Platelet count, ×109/l | ||
| <150,000 | 3 | 11 |
| ≥150,000 | 25 | 89 |
| β2M | ||
| <3 | 26 | 93 |
| >3 | 2 | 7 |
| Cytogenetics | ||
| 13q | 12 | 43 |
| Tri12 | 2 | 7 |
| Normal | 5 | 18 |
| 11q | 2 | 7 |
| 17p | 3 | 11 |
| Unknown | 4 | 14 |
| CD38 status | ||
| Positive | 6 | 21 |
| Negative | 20 | 72 |
| Unknown | 2 | 7 |
| ZAP-70 status | ||
| Positive | 1 | 3 |
| Negative | 7 | 25 |
| Unknown | 20 | 72 |
Comparing HCs with all patients with CLL/SLL
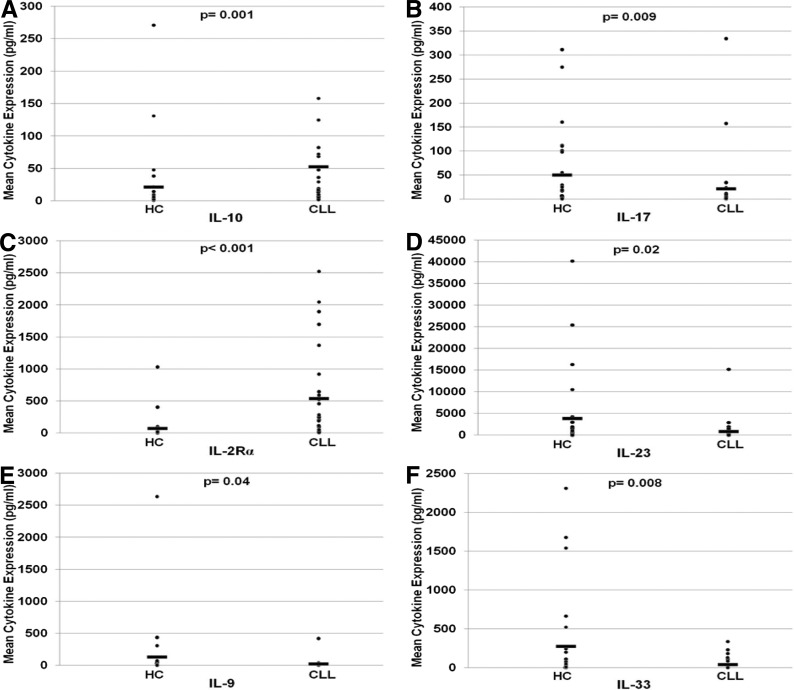
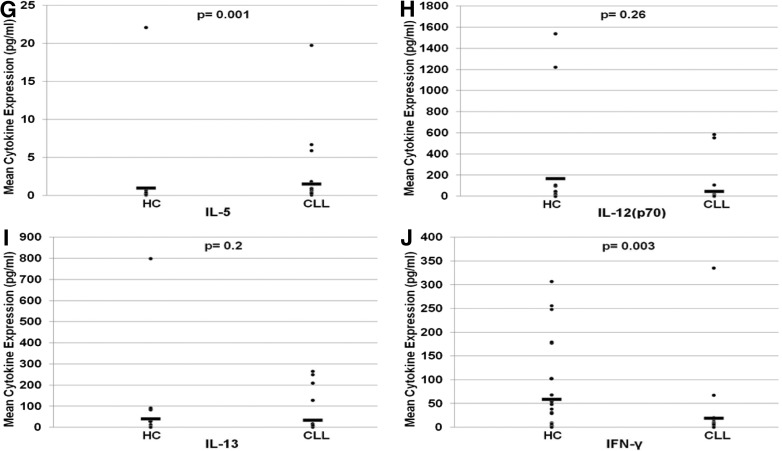
Differences in cytokine expression in CLL samples compared with HC samples are seen in Table 2. Statistically significant elevation in levels of Th2 cytokines IL-5 and IL-10 and depression in levels of Th1 cytokines IL-17, IL-23, and IFN-γ were noted. One exception to this pattern was a decrease in IL-33, a cytokine typically associated with a Th2 response. Increased expression of sIL-2Rα was most pronounced in the CLL/SLL population (8.99-fold; P<0.001), likely contributing to the predominance of Th2 immunity. As expected, β2M was also significantly higher in CLL/SLL patients compared with normal samples (1.79-fold; P=5.17×10−7). In univariate analyses, significantly higher expression in CLL/SLL patient serum compared with HCs was measured in the following chemokines: 6CKine, CTAK, I-309, MCP-1, MCP-4, MIP-1δ. Significantly lower expression in CLL/SLL patient serum compared with HCs was noted for ENA-78.
Table 2. Cytokine Expression Patterns and Their Relationship with Age, Sex, and Cytogenetic Profile in the CLL/SLL Population.
| Cytokine expresseda | Comparing healthy controls with CLL/SLL population as a whole |
Comparing healthy controls with CLL/SLL population stratified according to cytogenetics |
Analysis of CLL population only |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| HCs (n=28) |
All patients with CLL/SLL (n=28) |
P for means | FC in protein expression: HCs versus CLL |
GR CLL/SLL (n=12) |
FC in protein expression: HC versus GR CLL/SLL |
int/PR CLL/SLL (n=12) |
FC in protein expression: HCs versus int/PR CLL/SLL |
Multivariate analysis for mean protein expression in CLL/SLL population (P) |
|||
| Mean ± sem | Mean ± sem | FC | Mean | FC | Mean | FC | GR versus int/PR | Age | Sex | ||
| Cytokines with increased expression in the CLL/SLL population | |||||||||||
| β2M | 1.37 ± 0.1 | 2.7 ± 0.2 | <0.001 | 1.79 | 2.4 | NM | 3.0 | NM | 0.51 | 0.007 | 0.11 |
| 6CKine | 663.2 ± 83.2 | 2367.9 ± 544.3 | <0.001 | 2.78 | 2869.3 | 3.1 | 1844.6 | 3.2 | 0.51 | 0.51 | 0.88 |
| BCA1 | 34.0 ± 6.7 | 54.6 ± 6.0 | 0.001 | 1.72 | 48.9 | 1.4 | 60.7 | 2.2 | 0.35 | 0.39 | 0.91 |
| CTAK | 895.5 ± 96.8 | 1393.4 ± 96.5 | <0.001 | 1.6 | 1191.1 | 1.5 | 1604.5 | 1.6 | 0.04 | 0.85 | 0.58 |
| Eotaxin | 150.3 ± 29.0 | 173.9 ± 16.3 | 0.03 | 1.3 | 184.5 | 1.4 | 162.8 | 1.1 | 0.20 | 0.62 | 0.07 |
| Eotaxin2 | 939.7 ± 145.5 | 1542.8 ± 223.5 | 0.004 | 1.82 | 1572.2 | 1.6 | 1512.1 | 1.9 | 0.55 | 0.15 | 0.40 |
| Eotaxin 3 | 76.0 ± 28.0 | 159.2 ± 33.4 | 0.004 | 2.62 | 155.2 | 3.2 | 163.4 | 2.2 | 0.77 | 0.85 | 0.06 |
| FGF-2 | 60.9 ± 12.4 | 74.0 ± 12.9 | 0.9 | 1.13 | 55.0 | 1.3 | 93.9 | 1.7 | 0.19 | 0.06 | 0.15 |
| Flt3_Ligand | 46.9 ± 11.3 | 47.8 ± 10.7 | 0.6 | 1.5 | 59.9 | 2.0 | 35.1 | 1.0 | 0.05 | 0.81 | 0.007 |
| I-309 | 1.3 ± 0.1 | 2.8 ± 0.4 | 0.008 | 1.76 | 3.1 | 1.9 | 2.5 | 1.5 | 0.02 | 0.87 | 0.001 |
| IL-10 | 21.54 ± 10.5 | 29.5 ± 6.5 | 0.001 | 2.52 | 17.8 | 1.8 | 41.7 | 3.8 | 0.32 | 0.17 | 0.07 |
| IL-12(p40) | 30.67 ± 6.1 | 74.2 ± 34.8 | 0.82 | 1.02 | 117.5 | 1.4 | 29.1 | 1.3 | 0.10 | 0.51 | 0.14 |
| IL-13 | 12.3 ± 4.6 | 35.1 ± 14.3 | 0.20 | 1.4 | 52.0 | 1.4 | 17.5 | 1.6 | 0.25 | 0.92 | 0.97 |
| IL-15 | 5.25 ± 1.3 | 13.5 ± 7.8 | 0.82 | 1.07 | 20.5 | 1.2 | 6.2 | 1.1 | 0.16 | 0.39 | 0.08 |
| IL-16 | 85.44 ± 20.1 | 225.6 ± 41.1 | <0.001 | 3.98 | 242.7 | 3.3 | 207.7 | 5.3 | 1.00 | 0.97 | 0.40 |
| IL-1α | 100.87 ± 37.8 | 133.4 ± 31.9 | 0.6 | 1.28 | 179.4 | 1.8 | 85.4 | 1.3 | 0.06 | 0.05 | 0.48 |
| IL-1Rα | 19.58 ± 6.4 | 34.3 ± 25.1 | 1.0 | 1.09 | 57.4 | 1.0 | 10.3 | 1.0 | 0.17 | 0.43 | 0.11 |
| IL-28A | 10.49 ± 2.6 | 34.1 ± 8.6 | 0.001 | 2.68 | 45.4 | 2.9 | 22.4 | 3.1 | 0.27 | 0.19 | 0.73 |
| IL-5 | 0.98 ± 0.8 | 1.4 ± 0.5 | 0.001 | 2.24 | 0.9 | 2.7 | 1.9 | 2.2 | 0.44 | 0.54 | 0.67 |
| IP-10 | 296.42 ± 36.4 | 471.7 ± 77.3 | 0.26 | 1.19 | 417.7 | 1.1 | 528.1 | 1.4 | 0.58 | 0.57 | 0.85 |
| MCP-1 | 433.44 ± 37.7 | 1104.4 ± 164.6 | <0.001 | 2.1 | 1062.1 | 2.2 | 1148.5 | 2.3 | 0.69 | 0.48 | 0.96 |
| MCP-2 | 43.6 ± 2.9 | 52.6 ± 2.5 | 0.07 | 1.23 | 50.6 | 1.2 | 54.7 | 1.3 | 0.48 | 0.09 | 0.16 |
| MCP-4 | 125.4 ± 12.4 | 252.1 ± 21.9 | <0.001 | 2.0 | 249.2 | 2.1 | 255.1 | 2.2 | 0.83 | 0.29 | 0.92 |
| MIP-1δ | 3058.8 ± 544.5 | 4595.5 ± 489.2 | 0.002 | 1.55 | 3651.0 | 1.3 | 5581.1 | 2.2 | 0.02 | 0.81 | 0.83 |
| RANTES | 39,108.0 ± 9175.4 | 45,552.4 ± 9892.7 | 0.51 | NM | 29,181.7 | NM | 64,507.9 | NM | 0.24 | 0.79 | 0.12 |
| SCF | 7.08 ± 1.7 | 33.10 ± 4.4 | <0.001 | 5.42 | 25.3 | 3.2 | 41.3 | 8.3 | 0.20 | 0.41 | 0.004 |
| SDF-1α + β | 4122.0 ± 340.8 | 6214.5 ± 400.5 | 0.001 | 1.55 | 5541.7 | 1.3 | 6916.6 | 1.8 | 0.04 | 0.02 | 0.24 |
| sIL-2Rα | 71.11 ± 38.4 | 551.24 ± 117.3 | <0.001 | 8.99 | 258.6 | 2.8 | 856.6 | 33.9 | 0.006 | 0.67 | 0.51 |
| TARC | 96.8 ± 13.6 | 135.5 ± 20.5 | 0.28 | 116.8 | 1.5 | 155.0 | 1.2 | 0.51 | 0.38 | 0.28 | |
| TNF-α | 10.01 ± 2.6 | 25.35 ± 6.0 | 0.001 | 2.23 | 27.0 | 1.6 | 23.6 | 3.4 | 0.33 | 0.84 | 0.03 |
| TRAIL | 34.9 ± 3.6 | 51.9 ± 4.3 | 0.02 | 1.36 | 44.0 | 1.2 | 60.1 | 1.9 | 0.45 | 0.53 | 0.01 |
| Cytokines with decreased expression in the CLL/SLL population | |||||||||||
| EGF | 293.8 ± 133.1 | 142.2 ± 16.7 | 0.3 | 1.39 | 158.4 | 1.3 | 125.3 | 1.8 | 0.49 | 0.75 | 0.54 |
| ENA-78 | 1360.5 ± 283.7 | 761.1 ± 68.1 | 0.3 | 1.37 | 835.6 | 1.2 | 683.3 | 1.4 | 0.40 | 0.13 | 0.13 |
| Fractalkine | 425.2 ± 128.5 | 190.0 ± 74.0 | 0.006 | 5.4 | 217.3 | 5.6 | 161.5 | 4.9 | 0.33 | 0.78 | 0.08 |
| G-CSF | 56.2 ± 31.3 | 39.3 ± 5.4 | 0.02 | 1.29 | 49.5 | 1.6 | 28.7 | 1.0 | 0.006 | 0.74 | 0.19 |
| GM-CSF | 455.0 ± 177.3 | 105.1 ± 52.0 | 0.13 | 5.0 | 84.8 | 5.2 | 126.4 | 4.1 | 0.88 | 0.41 | 0.33 |
| GRO | 1537.8 ± 224.1 | 1403.1 ± 173.5 | 0.56 | 1.08 | 1230.4 | 1.1 | 1583.3 | 1.1 | 0.47 | 0.70 | 0.09 |
| IFN-α2 | 218.53 ± 83.0 | 162.3 ± 61.0 | 0.69 | 1.07 | 248.7 | 1.3 | 72.2 | 1.3 | 0.06 | 0.13 | 0.15 |
| IFN-γ | 59.07 ± 16.9 | 14.7 ± 7.3 | 0.003 | 5.65 | 10.5 | 5.9 | 19.1 | 3.9 | 0.76 | 0.36 | 0.42 |
| IL-12(p70) | 166.11 ± 85.2 | 43.6 ± 27.1 | 0.26 | 2.68 | 60.5 | 2.5 | 26.0 | 2.4 | 0.67 | 0.86 | 0.69 |
| IL-17 | 50.47 ± 15.4 | 19.5 ± 8.8 | 0.009 | 4.09 | 20.4 | 3.0 | 18.6 | 6.5 | 0.61 | 0.52 | 0.17 |
| IL-1β | 6.19 ± 5.1 | 3.8 ± 2.1 | 0.84 | 1.33 | 3.1 | 1.1 | 4.6 | 1.3 | 0.99 | 0.27 | 0.04 |
| IL-2 | 15.30 ± 8.8 | 14.0 ± 6.2 | 0.77 | 1.49 | 11.9 | 1.7 | 16.3 | 1.1 | 0.99 | 0.82 | 0.12 |
| IL-23 | 3898.7 ± 1728.4 | 1054.1 ± 478.2 | 0.02 | 5.19 | 1615.5 | 2.9 | 470.0 | 9.7 | 0.33 | 0.33 | 0.31 |
| IL-33 | 276.23 ± 110.8 | 48.4 ± 12.5 | 0.008 | 4.24 | 50.7 | 2.4 | 46.0 | 8.2 | 0.61 | 0.95 | 0.72 |
| IL-4 | 25.67 ± 19.9 | 23.2 ± 13.6 | 0.63 | 1.5 | 19.5 | 2.0 | 27.2 | 1.2 | 0.92 | 0.24 | 0.21 |
| IL-6 | 96.29 ± 49.9 | 33.9 ± 17.4 | 0.94 | 1.36 | 22.7 | 1.5 | 45.6 | 1.1 | 0.84 | 0.71 | 0.08 |
| IL-7 | 28.83 ± 13.1 | 21.0 ± 6.1 | 0.93 | 1.08 | 27.1 | 1.2 | 14.6 | 1.4 | 0.45 | 0.11 | 0.28 |
| IL-8 | 40.36 ± 9.3 | 36.4 ± 7.8 | 0.65 | 1.02 | 48.8 | 1.3 | 23.4 | 1.2 | 0.05 | 0.51 | 0.34 |
| IL-9 | 131.7 ± 94.6 | 23.2 ± 15.5 | 0.04 | 2.38 | 42.0 | 2.0 | 3.7 | 4.0 | 0.09 | 0.34 | 0.10 |
| MCP-3 | 26.1 ± 7.6 | 24.4 ± 5.7 | 0.27 | 1.28 | 33.8 | 1.1 | 14.5 | 1.6 | 0.07 | 0.12 | 0.89 |
| MDC | 2339.5 ± 254.7 | 2097.8 ± 141.4 | 0.77 | 1.05 | 2291.3 | 1.0 | 1895.9 | 1.1 | 0.22 | 0.71 | 0.91 |
| MIP-1α | 141.2 ± 33.0 | 76.0 ± 11.7 | 0.22 | 1.42 | 79.1 | 1.3 | 72.8 | 1.5 | 0.45 | 0.25 | 0.14 |
| MIP-1β | 156.5 ± 42.2 | 148.3 ± 28.0 | 0.25 | 1.27 | 151.0 | 1.2 | 145.5 | 1.5 | 0.56 | 0.56 | 0.04 |
| PDGF-AA | 27,114.5 ± 8093.9 | 15,757.1 ± 1374.0 | 0.07 | NM | 13,897.8 | NM | 17,910.1 | NM | 0.11 | 0.60 | 0.52 |
| PDGF-AA/BB | 47,959.1 ± 5232.0 | 41,567.1 ± 4106.4 | 0.2 | NM | 37,808.2 | NM | 45,919.6 | NM | 0.62 | 0.51 | 0.59 |
| TGF-α | 25.17 ± 5.0 | 12.73 ± 3.4 | <0.001 | 3.61 | 16.5 | 2.8 | 8.8 | 4.0 | 0.22 | 0.32 | 0.48 |
| TPO | 1658.6 ± 635.9 | 691.6 ± 120.5 | 0.53 | 1.53 | 584.9 | 1.1 | 802.8 | 2.3 | 0.49 | 0.20 | 0.88 |
| VEGF | 554.0 ± 97.7 | 344.7 ± 69.9 | 0.008 | 1.73 | 325.2 | 1.7 | 365.0 | 1.9 | 0.89 | 0.73 | 0.16 |
Fifty-nine cytokine/chemokines were measured in 28 patients with CLL/SLL. Expression patterns were mixed but skewed toward an increase in Th2 cytokines and a decrease in Th2/Th17/Th9 cytokines in the CLL/SLL population. NM, not measurable; BCA1, B cell-attracting chemokine 1; Flt3_Ligand, Fms-like tyrosine kinase 3 ligand; SCF, stem cell factor; SDF-1α + β, stromal cell-derived factor 1α + β; GRO, growth-regulated oncogene; MDC, macrophage-derived chemokine; PDGF, platelet-derived growth factor; TPO, thrombopoietin.
All cytokines in pg/ml, except for β2M, which is in μg/ml.
HCs were also compared with CLL/SLL patients stratified using a cytogenetics-based risk model (Table 2): CLL/SLL patients with GR cytogenetics (13q abnormalities) demonstrated a significant increase in Th2 cytokine expression, including IL-5 (2.7-fold; P=0.009) and IL-10 (1.8-fold; P=0.05), and a significant decrease Th1 cytokine IFN-γ (5.65-fold; P=0.009). sIL-2Rα trended to higher expression in the GR CLL population (P=0.19). Th2/Th1 dysregulation was amplified when comparing HCs with int/PR CLL/SLL patients (Fig. 1): a significant escalation in IL-10 and sIL-2Rα and de-escalation in IL-17, IL-23, and IFN-γ were found in int/PR CLL/SLL. Significant changes in IL-9, IL-33, I-309, and MIP-1δ were also noted. β2M was increased in int/PR patients when compared with GR patients (1.4-fold; P=0.035) and HCs (2.2-fold; P<0.001).
Figure 1. Comparison of cytokine expression concentration in HCs and CLL/SLL patients.
Dot-plots demonstrate differences in mean expression for IL-10 (A), IL-17 (B), sIL-2Rα(C), IL-23 (D), IL-9 (E), IL-33 (F), IL-5 (G), IL-12(p70), IL-12 (H), IL-13 (I), and IFN-γ (J) between HCs and CLL patients with statistical significance using Mann-Whitney analysis.
Comparing CLL/SLL patients with GR and int/PR cytogenetics
With the exception of sIL-2Rα, a multivariate analysis of the CLL population, including age, sex, and cytogenetic risk group, did not show significant changes in expression of cytokines, although changes did follow similar trends to those mentioned above (Table 2). Chemokines G-CSF, MIP-1δ, and I-309 maintained their significance in relation to cytogenetic risk without age or sex confounding results (P=0.006, P=0.02, and P=0.02, respectively). All three of these chemokines have been implicated in Th2 immunity. There was no significant difference in the absolute lymphocyte count between these two patient populations that could potentially contribute to a difference in cytokine expression. It is important to note that data from HCs were omitted in the multivariate analysis, as demographic information was not available in this population.
Use of a cytokine model to predict disease progression and need for therapy
Kruskal-Wallis analysis identified a number of cytokines/chemokines with significant differential expression among HCs, GR, and int/PR CLL/SLL (IL-5, IL-9, IL-10, IL-16, IL-17, IL-23, IL-28A, IL-33, I-309, IFN-γ, TGF-α, TNF-α, MIP-1δ, and sIL-2Rα). Significant deviations in expression for these specific cytokines, defined as >3 sem, were then determined. An IL-17/sIL-2Rα-based cytokine risk model was developed using these data (Table 3), as significant deviations in these two cytokines were found to have the best negative-predictive value for GR cytogenetics and positive-predictive value for both int/PR cytogenetics and need for therapy: among the patients GR cytogenetics, 33% of samples had a decrease in IL-17 and 30% had an increase in sIL-2R. These findings were exaggerated in the int/PR CLL/SLL population: 67% of samples had a decrease in IL-17, and 70% had an increase in sIL-2Rα. Fifteen patients in the CLL/SLL population as a whole showed a significant decrease in IL-17 and a significant increase in sIL-2Rα: 14 (99%) of these patients (13 CLL and one SLL) had int/PR cytogenetics, and one CLL patient had GR cytogenetics. Interestingly, this group also included all four patients who eventually received systemic therapy for disease (three patients with int/PR and one with GR cytogenetics).
Table 3. Decreased IL-17 and Increased IL-2Rα Predict for an int/PR or PR Cytogenetic Profile.
| Cytogenetic profile | IL-17 |
P | IL-2Rα |
P | ||||
|---|---|---|---|---|---|---|---|---|
| Mean ± sem | ≤3 sem; n = 17 [No. (%)] | >3 sem; n = 30 [No. (%)] | Mean ± sem | ≤3 sem; n = 24 [No. (%)] | >3 sem; n = 23 [No. (%)] | |||
| Healthy | 50.5 ± 15.3 | 71.1 ± 38.4 | ||||||
| GR CLL: 13q; n = 24 | 20.4 ± 10.4 | 14 (82%) | 10 (33%) | 258.6 ± 99.1 | 17 (71%) | 7 (30%) | ||
| int/PR CLL: Tri12, CN, 11q, or 17p; n = 23 | 19.0 ± 14.2 | 3 (18%) | 20 (67%) | 0.003 | 856.6 ± 199.4 | 7 (29%) | 16 (70%) | 0.006 |
More patients with int/PR cytogenetics demonstrated a significant decrease in IL-17 and increase in sIL-2Rα as compared with the GR CLL group, suggesting that the model combining IL-17/sIL-2Rα may predict for more aggressive disease. CN, cytogenetic normal.
DISCUSSION
Better prognostic/predictive markers in CLL/SLL will not only catch early disease progression but also potentially identify patients with early stage disease who may benefit from therapy at time of diagnosis. Moreover, tailoring therapy to such markers, by using available and novel targeted drugs, may improve survival outcomes. Given the large impact of the microenvironment on CLL/SLL survival, it is only reasonable to turn to the microenvironment to uncover such markers. This study comprehensively addresses whether cytokines in the CLL/SLL microenvironment may serve in this role in a unique manner. Similar studies have evaluated the correlation of a small panel of cytokines to biologic markers, including CD38, ZAP-70, and IgVH status [22, 23]. However, this study is the first to measure a large panel of 59 cytokines/chemokines and link patterns of expression to aggressive disease in the context of a cytogenetic-based risk model, a biologic model with the greatest clinical applicability at the current time.
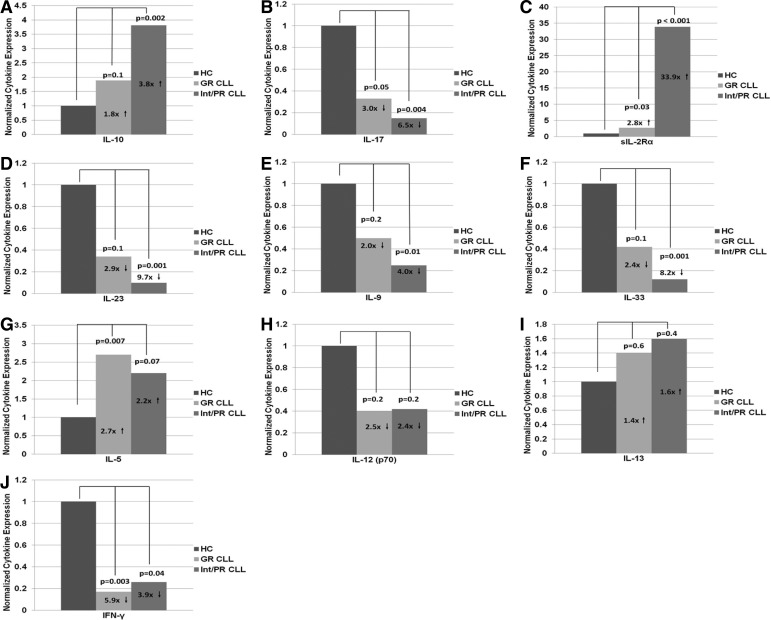
With the use of samples from HCs and patients with CLL/SLL, it was determined that the cytokine signature in the diseased population was somewhat mixed but tipped in favor of Th2 immunity, findings consistent with those described previously by other groups [11, 22, 24]. Increased expression in Th2 cytokines/chemokines (IL-5, IL-10, 1-309, MIP-1δ) and decreased expression in Th1/Th17 cytokines [IL-2, IL-12(p70), IL-17, IL-23, IFN-γ] were noted (Table 1 and Fig. 1). Cytokine/chemokine expression was subsequently compared between HCs and GR CLL/SLL and HCs and int/PR CLL/SLL—disease risk defined using cytogenetics. Th2 cytokine expression was amplified exponentially while Th1/Th17 cytokines decreased in a step-wise fashion when comparing HCs and GR and HCs and int/PR CLL/SLL (Fig. 2). This provides a mechanistic explanation for quick disease progression and poor survival outcomes associated with specific genotypic aberrations in CLL, namely, the increase in Th2 effect.
Figure 2. Changes in cytokine expression in HCs and CLL/SLL patients stratified according to cytogenetic risk.
FCs escalate from HCs to GR CLL/SLL to int/PR CLL/SLL for IL-10 (A) and sIL-2Rα (C) but de-escalate for IL-17 (B), IL-23 (D), IL-9 (E), and IL-33 (F) with statistical significance using two-sample t-tests. In the CLL population, a trend for escalation in IL-5 (G) and IL-13 (I) and a trend for de-escalation in IL-12(p70) (H) and IFN-γ (J) are also seen.
Of particular interest was the presence and degree of amplification in sIL-2Rα in HCs versus GR and HCs versus int/PR (Table 3) in this study. High levels of sIL-2Rα are known to have an immunomodulatory effect, reported to predict for an unfavorable prognosis in a variety of hematologic malignancies, including FL [25], cutaneous T cell lymphoma [26], hairy cell leukemia [27], and CLL [28]. The immunomodulatory role of sIL-2Rα appears to be pleiotropic depending on the disease state. In naïve FL treatment, Yang et al. [25] shed light on the “promoter effect” of sIL-2Rα, resulting in Stat5 signaling and proliferation in Foxp3-expressing Tregs. In the CLL population, the role of sIL-2Rα is less clear. With the use of data in this study, one could argue a similar mechanism to that seen in FL: sIL-2Rα likely serves as an inducer of IL-2-mediated Treg induction, leading to the abrogation of Th1, Th17, and potentially Th9 activity (a novel subset of Th cells that secretes IL-9) [29–31], demonstrable by a decrease in cytokines produced by these specific Th subsets (Fig. 3). This may have led to the unopposed Th2 effect with subsequent increase in Th2 cytokines/chemokines seen on our CLL/SLL population (Fig. 2).
Figure 3. Proposed mechanisms of CLL immunity.
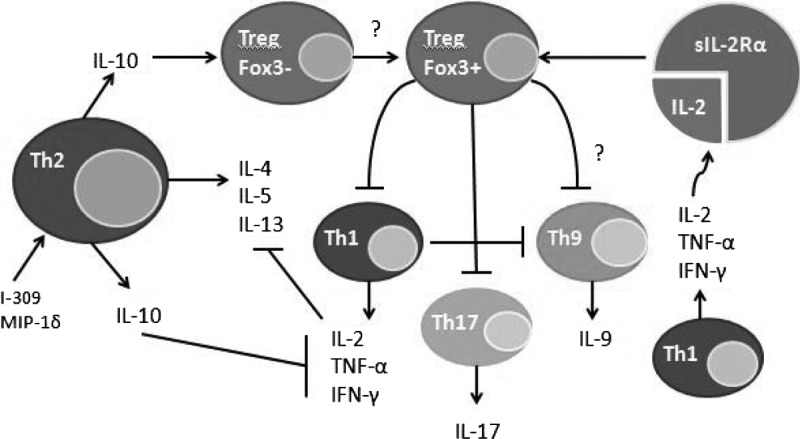
Up-regulation of Tregs may suppress other Th subsets, including Th1, Th17, and Th9 T cells, leading to an unopposed increase in Th2 T cells. Our data are consistent with this mechanism. This anti-inflammatory response results in immune evasion of B-CLL cells [25, 29, 31].
Among the 59 cytokines/chemokines that were evaluated, we created a model using sIL-2Rα and IL-17, which correlated best with the cytogenetic profile. Similar to a model described by Tefferi et al. [32] for primary myelofibrosis, our model predicted a need for treatment as well—serum from all four patients requiring therapy for disease progression demonstrated IL-17 levels at least 3 sem below mean levels measured in HCs and sIL-2Rα levels at least 3 sem above mean levels measured in HCs, irrespective of their baseline genetic profile (Table 3). This model may prove useful in early detection of progression or even relapse post-therapy in the absence of clinical symptoms. Continued follow-up of our current sample population and future studies in a larger population in need of treatment are warranted to validate our findings.
A limitation to this study is that the HCs were not age-matched to a CLL/SLL population younger than the typical demographic. Our small sample size also made it difficult to determine whether one specific cytogenetic abnormality contributed to our findings more than another. In fact, we suspect that this Th2/Th1 skew may not be as pronounced in patients with 17p deletion, which may explain the efficacy of alemtuzumab—a drug known to suppress Th1 subsets—in CLL/SLL patients with this chromosomal abnormality [33].
Despite the limitations of this study, findings provide insight into the pathogenesis of CLL/SLL in the context of a chromosomal-based prognostic model. Moreover, they help to explain why certain anti-CLL pharmacological agents may be effective. For example, patients with higher levels of IL-10 may benefit from the use of BTK inhibitors, as BTK PC132765 has been shown to decrease IL-10 levels, a change that may potentially contribute to its efficacy [34]. Similarly, in a study with lenalidomide, Ferrajoli et al. [24] showed that responders to lenalidomide had lower levels of IL-10 and sIL-2Rα with increased levels of IFN-γ, implying that lenalidomide exerts an immune-modulatory effect that favors Th1 activity. Taking things one step further, findings in this study support the concept of individualized therapy. They also rationalize the testing of novel targeted drugs aimed at the cytokine milieu in CLL/SLL—daclizumab, a mAb that blocks sIL-2Rα activity is one potential candidate [35]. Taken collectively, it is clear that studies aimed at deconstructing the tumor microenvironment are needed to identify better prognostic/predictive biomarkers that may ideally dictate therapy with the ultimate goal of impacting survival.
ACKNOWLEDGMENTS
This study was primarily supported by a private philanthropic donation through Rush University Medical Center. In addition, K.W.C. was supported during this time period by the Leukemia and Lymphoma Society (6044-08), the American Association for Cancer Research (07-10-19-CHRI), the National Blood Foundation/American Association of Blood Banks (031824), the U.S. National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases award (DK074892), the Rubschlager Foundation, and the Coleman Foundation. We thank Stephen Rundlett for his assistance in analyzing the Milliplex data, Drs. Youping Deng and Sanjib Basu for their statistical support, and Elizabeth Barraza for her involvement in editing the manuscript.
Footnotes
- 6CKine
- 11q
- deletion 11q22-23
- 13q
- deletion 13q(14.3)
- 17p
- deletion 17p13 (p53)
- β2M
- β 2-microglobulin
- BTK
- Bruton's tyrosine kinase
- CLL
- chronic lymphocytic leukemia
- ENA-78
- epithelial neutrophil-activating protein 78
- FC
- fold-change
- FISH
- fluorescence in situ hybridization
- FL
- follicular lymphoma
- Foxp3
- forkhead box p3
- GR
- good risk
- HC
- healthy control
- IgVH
- Ig heavy chain variable gene region
- int/PR
- intermediate/poor risk
- IP-10
- IFN-γ-inducible protein
- PR
- poor risk
- s
- soluble
- SLL
- small lymphocytic leukemia
- Treg
- T regulatory cell
- Tri12
- trisomy 12
AUTHORSHIP
K.W.C. and L.A.P. conceived of and designed the research project. R.R.F., S.J., and L.A.P. designed and performed research experiments. R.R.F., L.A.P., R.K., K.W.C., M.L.L., P.V., and S.A.G. analyzed and interpreted data. R.K., L.A.P., and K.W.C. wrote the manuscript.
DISCLOSURES
The authors declare no competing financial interests.
REFERENCES
- 1. Muzio M., Bertilaccio M. T., Simonetti G., Frenquelli M., Caligaris-Cappio F. (2009) The role of Toll-like receptors in chronic B-cell malignancies. Leuk. Lymphoma 50, 1573–1580 [DOI] [PubMed] [Google Scholar]
- 2. Chiorazzi N., Rai K. R., Ferrarini M. (2005) Chronic lymphocytic leukemia. N. Engl. J. Med. 352, 804–815 [DOI] [PubMed] [Google Scholar]
- 3. Stevenson F. K., Caligaris-Cappio F. (2004) Chronic lymphocytic leukemia: revelations from the B-cell receptor. Blood 103, 4389–4395 [DOI] [PubMed] [Google Scholar]
- 4. Ghia P., Caligaris-Cappio F. (2006) The origin of B-cell chronic lymphocytic leukemia. Semin. Oncol. 33, 150–156 [DOI] [PubMed] [Google Scholar]
- 5. Seiffert M., Dietrich S., Jethwa A. (2012) Exploiting biological diversity and genomic aberrations in chronic lymphocytic leukemia. Leuk. Lymphoma 53, 1023–1031 [DOI] [PubMed] [Google Scholar]
- 6. Lagneaux L., Delforge A., Bron D., De Bruyn C., Stryckmans P. (1998) Chronic lymphocytic leukemic B cells but not normal B cells are rescued from apoptosis by contact with normal bone marrow stromal cells. Blood 91, 2387–2396 [PubMed] [Google Scholar]
- 7. Burger J. A., Tsukada N., Burger M., Zvaifler N. J., Dell'Aquila M., Kipps T. J. (2000) Blood-derived nurse-like cells protect chronic lymphocytic leukemia B cells from spontaneous apoptosis through stromal cell-derived factor-1. Blood 96, 2655–2663 [PubMed] [Google Scholar]
- 8. Tsukada N., Burger J. A., Zvaifler N. J., Kipps T. J. (2002) Distinctive features of “nurselike” cells that differentiate in the context of chronic lymphocytic leukemia. Blood 99, 1030–1037 [DOI] [PubMed] [Google Scholar]
- 9. Gallego A., Vargas J. A., Castejon R., Citores M. J., Romero Y., Millán I., Durántez A. (2003) Production of intracellular IL-2, TNF-α, and IFN-γ by T cells in B-CLL. Cytometry B Clin. Cytom. 56, 23–29 [DOI] [PubMed] [Google Scholar]
- 10. Burger J. A. (2011) Nurture versus nature: the microenvironment in chronic lymphocytic leukemia. Hematology Am. Soc. Hematol. Educ. Program 2011, 96–103 [DOI] [PubMed] [Google Scholar]
- 11. Christopoulos P., Pfeifer D., Bartholome K., Follo M., Timmer J., Fisch P., Veelken H. (2011) Definition and characterization of the systemic T-cell dysregulation in untreated indolent B-cell lymphoma and very early CLL. Blood 117, 3836–3846 [DOI] [PubMed] [Google Scholar]
- 12. Damle R. N., Wasil T., Fais F., Ghiotto F., Valetto A., Allen S. L., Buchbinder A., Budman D., Dittmar K., Kolitz J., Lichtman S. M., Schulman P., Vinciguerra V. P., Rai K. R., Ferrarini M., Chiorazzi N. (1999) Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood 94, 1840–1847 [PubMed] [Google Scholar]
- 13. Hamblin T. J., Davis Z., Gardiner A., Oscier D. G., Stevenson F. K. (1999) Unmutated IgVH genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood 94, 1848–1854 [PubMed] [Google Scholar]
- 14. Ferrarini M., Chiorazzi N. (2004) Recent advances in the molecular biology and immunobiology of chronic lymphocytic leukemia. Semin. Hematol. 41, 207–223 [DOI] [PubMed] [Google Scholar]
- 15. Chen L., Widhopf G., Huynh L., Rassenti L., Rai K. R., Weiss A., Kipps T. J. (2002) Expression of ZAP-70 is associated with increased B-cell receptor signaling in chronic lymphocytic leukemia. Blood 100, 4609–4614 [DOI] [PubMed] [Google Scholar]
- 16. Döhner H., Stilgenbauer S., Benner A., Leupolt E., Kröber A., Bullinger L., Döhner K., Bentz M., Lichter P. (2000) Genomic aberrations and survival in chronic lymphocytic leukemia. N. Engl. J. Med. 343, 1910–1916 [DOI] [PubMed] [Google Scholar]
- 17. Döhner H., Stilgenbauer S., Döhner K., Bentz M., Lichter P. (1999) Chromosome aberrations in B-cell chronic lymphocytic leukemia: reassessment based on molecular cytogenetic analysis. J. Mol. Med. (Berl). 77, 266–281 [DOI] [PubMed] [Google Scholar]
- 18. Neilson J. R., Auer R., White D., Bienz N., Waters J. J., Whittaker J. A., Milligan D. W., Fegan C. D. (1997) Deletions at 11q identify a subset of patients with typical CLL who show consistent disease progression and reduced survival. Leukemia 11, 1929–1932 [DOI] [PubMed] [Google Scholar]
- 19. Zenz T., Mohr J., Edelmann J., Sarno A., Hoth P., Heuberger M., Helfrich H., Mertens D., Dohner H., Stilgenbauer S. (2009) Treatment resistance in chronic lymphocytic leukemia: the role of the p53 pathway. Leuk. Lymphoma 50, 510–513 [DOI] [PubMed] [Google Scholar]
- 20. Stilgenbauer S., Sander S., Bullinger L., Benner A., Leupolt E., Winkler D., Kröber A., Kienle D., Lichter P., Döhner H. (2007) Clonal evolution in chronic lymphocytic leukemia: acquisition of high-risk genomic aberrations associated with unmutated VH, resistance to therapy, and short survival. Haematologica 92, 1242–1245 [DOI] [PubMed] [Google Scholar]
- 21. Geisler C., Jurlander J., Bullinger L., Sander S., Brown P., Benner A., Philip P., Döhner H., Stilgenbauer S. (2009) Danish CLL2-study revisited: FISH on a cohort with a 20-yr follow-up confirms the validity of the hierarchical model of genomic aberrations in chronic lymphocytic leukaemia. Eur. J. Haematol. 83, 156–158 [DOI] [PubMed] [Google Scholar]
- 22. Yan X. J., Dozmorov I., Li W., Yancopoulos S., Sison C., Centola M., Jain P., Allen S. L., Kolitz J. E., Rai K. R., Chiorazzi N., Sherry B. (2011) Identification of outcome-correlated cytokine clusters in chronic lymphocytic leukemia. Blood 118, 5201–5210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kara I. O., Sahin B., Gunesacar R. (2007) Expression of soluble CD27 and interleukins-8 and -10 in B-cell chronic lymphocytic leukemia: correlation with disease stage and prognosis. Adv. Ther. 24, 29–40 [DOI] [PubMed] [Google Scholar]
- 24. Ferrajoli A., Lee B. N., Schlette E. J., O'Brien S. M., Gao H., Wen S., Wierda W. G., Estrov Z., Faderl S., Cohen E. N., Li C., Reuben J. M., Keating M. J. (2008) Lenalidomide induces complete and partial remissions in patients with relapsed and refractory chronic lymphocytic leukemia. Blood 111, 5291–5297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang Z. Z., Grote D. M., Ziesmer S. C., Manske M. K., Witzig T. E., Novak A. J., Ansell S. M. (2011) Soluble IL-2Rα facilitates IL-2-mediated immune responses and predicts reduced survival in follicular B-cell non-Hodgkin lymphoma. Blood 118, 2809–2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dummer R., Posseckert G., Nestle F., Witzgall R., Burger M., Becker J. C., Schäfer E., Wiede J., Sebald W., Burg G. (1992) Soluble interleukin-2 receptors inhibit interleukin 2-dependent proliferation and cytotoxicity: explanation for diminished natural killer cell activity in cutaneous T-cell lymphomas in vivo? J. Invest. Dermatol. 98, 50–54 [DOI] [PubMed] [Google Scholar]
- 27. Chilosi M., Semenzato G., Cetto G., Ambrosetti A., Fiore-Donati L., Perona G., Berton G., Lestani M., Scarpa A., Agostini C.., et al. (1987) Soluble interleukin-2 receptors in the sera of patients with hairy cell leukemia: relationship with the effect of recombinant α-interferon therapy on clinical parameters and natural killer in vitro activity. Blood 70, 1530–1535 [PubMed] [Google Scholar]
- 28. Foa R., Fierro M. T., Giovanelli M., Lusso P., Benetton G., Bonferroni M., Forni G. (1987) Immunoregulatory T-cell defects in B-cell chronic lymphocytic leukemia: cause or consequence of the disease? The contributory role of decreased availability of interleukin-2 (IL-2). Blood Cells 12, 399–412 [PubMed] [Google Scholar]
- 29. Wong M. T., Ye J. J., Alonso M. N., Landrigan A., Cheung R. K., Engleman E., Utz P. J. (2010) Regulation of human Th9 differentiation by type I interferons and IL-21. Immunol. Cell Biol. 88, 624–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Goswami R., Kaplan M. H. (2011) A brief history of IL-9. J. Immunol. 186, 3283–3288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Romagnani S. (2002) Cytokines and chemoattractants in allergic inflammation. Mol. Immunol. 38, 881–885 [DOI] [PubMed] [Google Scholar]
- 32. Tefferi A., Vaidya R., Caramazza D., Finke C., Lasho T., Pardanani A. (2011) Circulating interleukin (IL)-8, IL-2R, IL-12, and IL-15 levels are independently prognostic in primary myelofibrosis: a comprehensive cytokine profiling study. J. Clin. Oncol. 29, 1356–1363 [DOI] [PubMed] [Google Scholar]
- 33. Kiaii S., Choudhury A., Mozaffari F., Rezvany R., Lundin J., Mellstedt H., Osterborg A. (2006) Signaling molecules and cytokine production in T cells of patients with B-cell chronic lymphocytic leukemia: long-term effects of fludarabine and alemtuzumab treatment. Leuk. Lymphoma 47, 1229–1238 [DOI] [PubMed] [Google Scholar]
- 34. Herman S. E., Gordon A. L., Hertlein E., Ramanunni A., Zhang X., Jaglowski S., Flynn J., Jones J., Blum K. A., Buggy J. J., Hamdy A., Johnson A. J., Byrd J. C. (2011) Bruton tyrosine kinase represents a promising therapeutic target for treatment of chronic lymphocytic leukemia and is effectively targeted by PCI-32765. Blood 117, 6287–6296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Matutes E. (2012) Novel and emerging drugs for rarer chronic lymphoid leukaemias. Curr. Cancer Drug Targets 12, 484–504 [DOI] [PubMed] [Google Scholar]